2025 DOCK tutorial 2 with PDBID 1XMU
Contents
Introduction
Structure Preparation
The objectives of this section are:
1. Structural Evaluation
a. Identify any missing loops in the protein structure b. Assess metal coordination atoms for key interactions with the protein
2. Protein Receptor Preparation
a. Generate protein structure in mol2 format b. Add hydrogen atoms c. Assign appropriate charges d. Save the refined protein structure in mol2 format
3. Ligand Preparation
a. Generate ligand structure in mol2 format b. Add hydrogen atoms c. Assign appropriate charges d. Save the refined ligand structure in mol2 format
1. Structural Evaluation
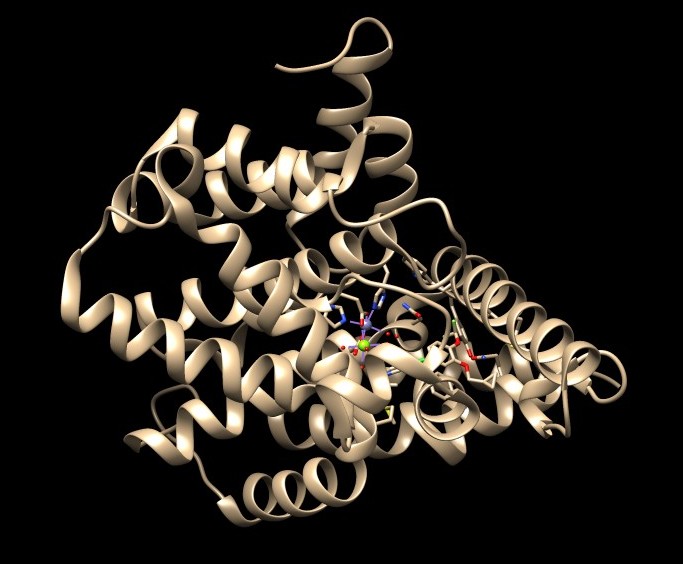
a. Open the downloaded PDB structure in Chimera (In this tutorial, 1XMU). Check for missing loops—these appear as dashed lines in the structure.
b. There are no missing loops in the structure 1XMU, so no further action is needed.
If you find missing loops in your structure, you will need to fix them before proceeding. Refer to (Tutorial 2025 [[1]]) for instructions on fixing missing loops.
b. In 1XMU, there is metal coordination atoms (Zinc and Magnesium) interacting with the protein. It is important to keep them intact and not delete them during protein preparation
2. Protein Receptor Preparation
a. Generate protein structure in MOL2 format
i. Open the downloaded pdb file (1XMU) in Chimera,
ii. Remove ligand
Go to Select → residue → ligand to highlight the ligand. Then go to Actions → Atoms/Bonds → Delete
iii. Repeat the process to delete water molecules by selecting HOH instead of ligand
iv. Save the protein, Go to File → Save Mol2. 1XMU_Rec_nCH.mol2 ('no' Charge and Hydrogen)
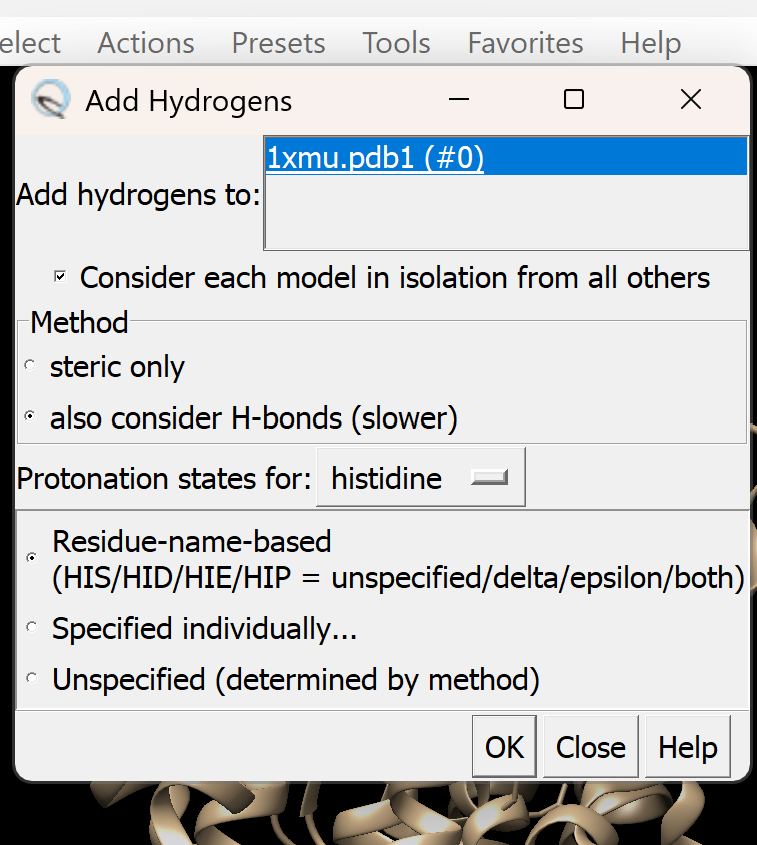
b. Add Hydrogen atoms
i. Go to Tools → Structure Editing → AddH. A dialogue box like this will appear, adjust as needed
ii. Select OK to proceed. If successful, You will get a confirmation message 'Hydrogens Added'
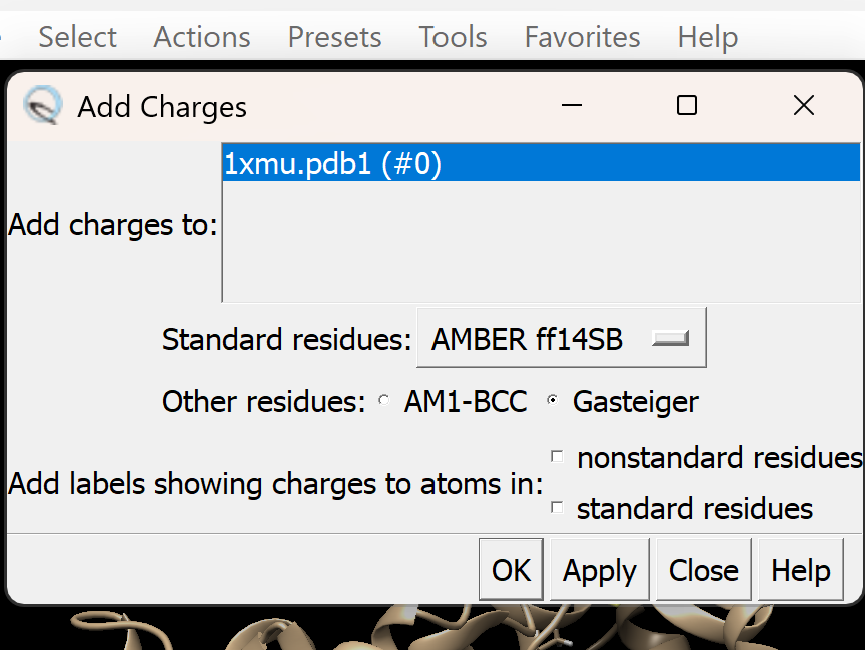
c. Assign appropriate charges
i. Go to Tools → Structure Editing → Add Charge.
A dialogue box like this will appear, adjust as needed. For this tutorial, we will use Gasteiger charges. Leave all other settings as they are, then click OK.
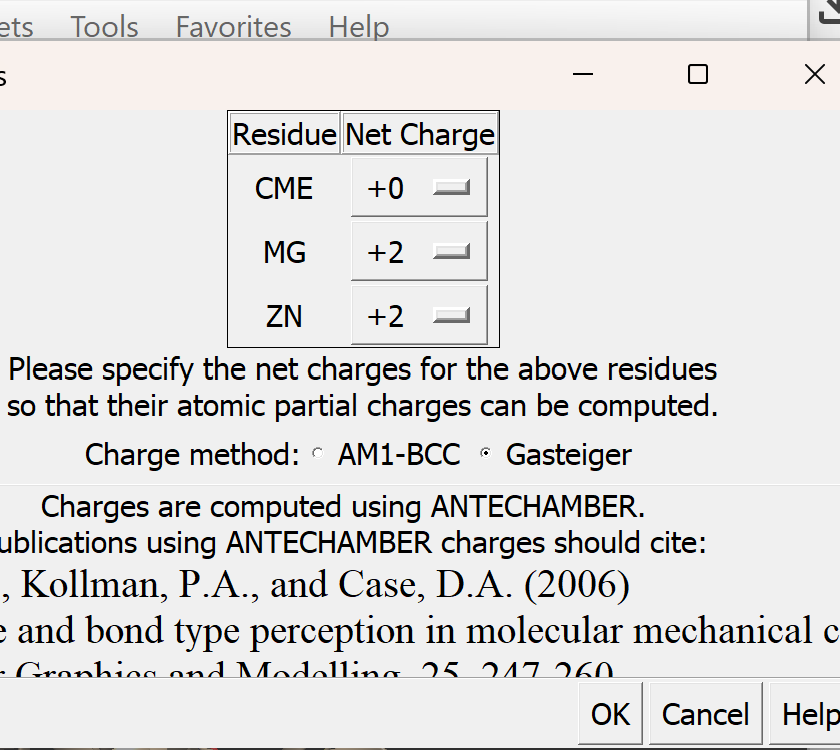
ii. A new pop-up menu will appear
iii. Just as before, make sure that Gasteiger is selected. All other defaults are okay.
iv. Select OK. If successful, You will get a confirmation message 'standard charges added'
d. Go to File → Save Mol2. 1XMU_Rec_wCH.mol2 ('with' Charge and Hydrogen)
3. Ligand Preparation