2018 DOCK tutorial 2 with PDBID 1C87
For additional Rizzo Lab tutorials see DOCK Tutorials. Use this link Wiki Formatting as a reference for editing the wiki. This tutorial was developed collaboratively by the AMS 536 class of 2018, using DOCK v6.8 and it shows how to dock a ligand into a receptor.
Contents
I. Introduction
DOCK
DOCK is a molecular docking program used in drug discovery. It was developed by Irwin D. Kuntz, Jr. and colleagues at UCSF (see UCSF DOCK). This program, given a protein binding site and a small molecule, tries to predict the correct binding mode of the small molecule in the binding site, and the associated binding energy. Small molecules with highly favorable binding energies could be new drug leads. This makes DOCK a valuable drug discovery tool. DOCK is typically used to screen massive libraries of millions of compounds against a protein to isolate potential drug leads. These leads are then further studied, and could eventually result in a new, marketable drug. DOCK works well as a screening procedure for generating leads, but is not currently as useful for optimization of those leads.
DOCK 6 uses an incremental construction algorithm called anchor and grow. It is described by a three-step process:
- Rigid portion of ligand (anchor) is docked by geometric methods.
- Non-rigid segments added in layers; energy minimized.
- The resulting configurations are 'pruned' and energy re-minimized, yielding the docked configurations.
1C87
In this tutorial we will use PDB code 1C87, which is the crystal structure of protein tyrosine phosphatase 1B complexed with 2-(oxalyl-amino-4,7-dihydro-5H-thieno[2,3-C]pyran-3-carboxylic acid.
Organizing Directories
We are going to create and organize directories so it would be easier for us to find or identify files in each directory. The structure of your directories should look something like this:
~/gpfs/projects/AMS536/2018/
/001.files/
/002.surface-spheres/
/003.box/
/004.grid/
/005.dock/
/006.virtual-screen/
|
To locate which directory you are currently in, type pwd. From your personal directory (...../AMS536/2018/yourName/), you can use the mkdir command to make directories. One way to make a number of directories is:
mkdir 001.files 002.spheres 003.box 004.grid 005.dock 006.virtual-screen
To see if you were successful, type
ls -l
to list directory contents. If you make a mistake, you can change the name of the directory or remove it and try again
to rename a directory..
mv oldname/ newname/
to remove a directory (must be empty)
rmdir mydirectory/
II. Preparing the Receptor and Ligand
Download the PDB File (1C87)
We are going to the PDB website (https://www.rcsb.org/) to download 1C87.pdb file and transfer this pdb file to your directory. First, open Chimera and load 1C87.pdb file. For dock, it is important that the receptor is separate entity and saved as its own structure file. We must also think about what parts of the structure should be included in the receptor and what parts should be ignored. (We may want some crystallographic water which interact with the binding site/ligand but may choose to remove other surrounding waters further away. Remove the ligand and save the receptor in mol2.format. "mol2" format shows the bond types (whether it is single or double bond), and the connectivity of the atoms (what is bonded to what). After saving the receptor with no Hydrogen (noh_receptor_1c87.mol2) and the receptor with built Hydrogens and partial charges (rec_withH_charge_1c87.mol2), load the original 1c87.pdb again. Remove the receptor and water, and save the ligand with no Hydrogen (noh_lig_1c87.mol2) and ligand with Hydrogen and partial charges (lig_withH_charge_1c87.mol2). In order to build Hydrogens and add charge on these structures, go to Tools and Structure Editing and use Dockprep. Along with building Hydrogens and adding partial charges, dockprep will try to fill in missing info from the pdb and also save the structures in a mol2 format at the end . (NOTE: For this system, when you add H on ligand, make the charge -1. In other cases, check the article that is related to pdb file to decide how it should be pronated or depronated.)
Go to
Tools -> Structure Editing -> Dockprep
Check the appropriate boxes:
(### Need photo of dockprep setup ###)
Or more explicitly:
Tools -> Structure Editing -> Add H Tools -> Structure Editing-> Add Charge Select AMBER ff99SB for Standard Residues, and add a charge of -1. File -> Save Mol2... -> lig_withH_charge.mol2
Lastly, open the receptor file and tell chimera to show the surface of your receptor. After this is calculated and you can see the surface of the protein, go to Structure Editing and write DMS file. Make sure to save all files. So far, we will have these files ready.
<pre>
1c87.pdb lig_withH_charge_1c87.mol2 noh_receptor_1c87.mol2 noh_lig_1c87.mol2 rec_withH_1c87.mol2 rec_1c87.dms |
After saving these two files with H, transfer files into 001.files directory:
scp -r ./*1c87* username@login.seawulf.stonybrook.edu:/gpfs/projects/AMS536/2018/your_directory_name/001.files
III. Generating Receptor Surface and Spheres
Generating the Receptor Surface
Make sure sphere directory is created and open the directory:
mkdir 002.surface-spheres cd 002.surface-spheres
Open Chimera and load the receptor surface:
Load the receptor (noh_receptor_1c87.mol2) file, which is located in the 001.files directory. Go to Action -> Surface -> Show and it will show the surface receptor. Next, save a DMS file as noh_surface_rec.dms by clicking Tools -> Structure editing -> Write DMS.
Creating Spheres We are going to create spheres by using the Sphgen program, which is a sphere generation program in Dock.
1. Create an input file
vi INSPH
Copy this following script in INSPH input file.
./noh_surface_rec.dms R X 0.0 4.0 1.4 1c87.spheres.sph
The first line ./noh_surface_rec.dms specifies the input file. R means that spheres generated will be outside of the receptor surface. X specifies all the points will be used. 0.0 is the distance in angstrom and it will avoid steric clashes. 4.0 is the maximum surface radius of the spheres and 1.4 is the minimum radius in angstroms.The last line 1c87.spheres.sph creates the file that has clustered spheres.
2. Run the Sphgen program
sphgen -i INSPH -o OUTSPH
-i chooses input file and -o writes toutput file.
3. Open Chimera to visualize the generated spheres Load 1c87.recep.sph file.
Now, we are going to select the spheres that is close to the native ligand molecule, which would be within 8.0 angstroms of the ligand.
sphere_selector 1c87.recep.sph ../001.files/lig_withH_charge_1c87.mol2 8.0
1c87.recep.pdbwill be created and load this file in Chimera.
IV. Generating Box and Grid
Creating the box
Enter 003.box-grid directory. Create input showbox.in:
Y 8.0 ../002.spheres/selected_spheres.sph 1 1c87.box.pdb
The first line Y creates a box (grid). 8.0 is the box length in angstroms. 1 means spheres. The last line, 1c87.box.pdb, outputs the box to the file.
Use this input type:
showbox < showbox.in
Now we can visualize the box in Chimera. Load 1c87.box.pdb.
Calculating Energy Grid
Now, we will compute the energy grid. Create grid.in file This saves the interaction energy of the receptor at each grid point
vi grid.in
Save the output in the file grid.out.
grid -i grid.in > grid.out
Answer the questions:
compute_grids yes grid_spacing 0.4 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4 bump_filter yes bump_overlap 0.75 receptor_file ../001.files/rec_withH_charge_1c87.mol2 box_file ../003.box/1c87.box.pdb vdw_definition_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
Once completed to answer the questions, it will create two output files: grid.bmp and grid.nrg
V. Docking a Single Molecule for Pose Reproduction
We would like to re-dock 1C87 into a receptor by using these methods: rigid docking, flex docking and fixed anchor docking.
Minimization
First, we are going to run minimization. Minimization is important because the structure of the ligand in the crystal may not be suited for the forcefield used in our calculations. Minimization relaxes the structure to the forcefield so that contact geometries are at an energy minimum. Create min.in file and run dock with this input file.
touch min.in ../../../zzz.programs/dock6/bin/dock6 -i min.in
Answer the questions like we did above:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../001.files/lig_withH_charge_1c87.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../001.files/lig_withH_charge_1c87.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary yes continuous_score_secondary no cont_score_rec_filename ../001.files/rec_withH_charge_1c87.mol2 cont_score_att_exp 6 cont_score_rep_exp 12 cont_score_rep_rad_scale 1 cont_score_use_dist_dep_dielectric yes cont_score_dielectric 4.0 cont_score_vdw_scale 1 cont_score_es_scale 1 footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix ligand_outfile_prefix 1c87.min write_orientations no num_scored_conformers 1 rank_ligands no
Now, 1c87.min_scored.mol2 will be produced. We can compare minimized ligand structure with unminimized ligand structure.
Rigid Docking
We are going to run rigid docking. With rigid docking the internal degrees of freedom of the ligand are constrained, and only translation and rotation of the whole structure is considered for docking. In order to run dock, we will have to create rigid.in file.
touch rigid.in ../../../zzz.programs/dock6/bin/dock6 -i rigid.in
Answer the questions:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../001.files/lig_withH_charge_1c87.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../001.files/lig_withH_charge_1c87.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../004.grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 1c87.rigid_restraint write_orientations no num_scored_conformers 40 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
This will create 1c87.rigid_restraint_scored.mol2 in 005.dock directory.
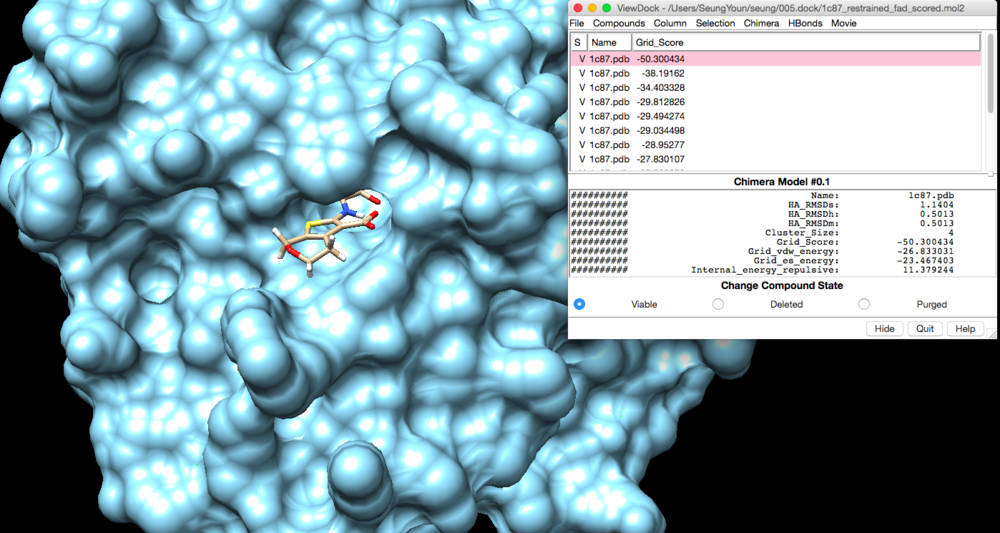
We can view rigid docking output file in CHIMERA.
Tools --> Surface/Binding Analysis -> ViewDock Select the output file -> select Dock 4,5, or 6
We can view the lowest grid score (the best ligand that fits in the pocket) , which is what we want, by clicking Column -> Show -> Grid_Score.
Flexible Docking
In order to run flexible docking, we will also create flex.in file. In this case, ligand and internal angles are flexible (NOTE: orient_ligand is set to yes).
touch flex.in ../../../zzz.programs/dock6/bin/dock6 -i flex.in
When prompted, answer the questions as follows
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../001.files/lig_withH_charge_1c87.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../001.files/lig_withH_charge_1c87.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../004.grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 1c87_flex write_orientations no num_scored_conformers 40 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
The output file is 3pgl.flex_scored.mol2, and is saved in the directory from where you ran dock. Then we can visualize the flexible docking in Chimera:
Fixed Anchor Docking
We are going to run fixed anchor docking. Create fad.in.The central ligand structure is fixed in the active site and internal angles are flexible and sampled. (NOTE: orient_ligand is set to no).
touch fad.in ../../../zzz.programs/dock6/bin/dock6 -i fad.in
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../001.files/lig_withH_charge_1c87.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../001.files/lig_withH_charge_1c87.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../004.grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 1c87_fad write_orientations no num_scored_conformers 40 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
Again, we can view fixed anchor docking in Chimera and find the lowest grid score value.
Now we have created output files for rigid, flexible and fixed anchor docking.
Molecular Footprint
When it comes to examining how a ligand interacts with its receptor, we can break it down to the interaction contribution of side chains in the binding pocket. Some interactions will be much stronger than other, with some being strongly favorable or strongly prohibitive to binding. Typically we want to look at those residues which contribute most to the interaction between ligand and receptor, favorable or not, so that we can deduce ideal physiochemical structures for future lead refinement. Dock can show this in what is called a Footprint. The plot shows electrostatic and van der Waals interaction contribution for the top X highest residue interactions vs the contribution of another ligand (In this case, we want to see the footprint of a given ligand with respect to the known binder ligand 1c87 from either its crystal structure, or its minimized structure). Here we will compare the footprint of minimized 1c87 vs its crystal structure.
Create a new directory for the footprint calculations, and create the input file for dock inside that directory:
mkdir 007.footprint cd !$ touch footprint.in
Answer the questions to fill in the input parameters like before:
conformer_search_type rigid use_internal_energy no ligand_atom_file ../001.files/1c87.min.mol2 ### NOTE: This can be changes based on what you want to compare with what ### limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary yes footprint_similarity_score_secondary no fps_use_footprint_reference_mol2 yes fps_footprint_reference_mol2_filename ../001.files/lig_withH_charge_1c87.mol2 ### Reference ligand for comparison of footprints ### fps_foot_compare_type Euclidean fps_normalize_foot no fps_foot_comp_all_residue yes fps_receptor_filename ../001.files/rec_withH_1c87.mol2 fps_vdw_att_exp 6 fps_vdw_rep_exp 12 fps_vdw_rep_rad_scale 1 fps_use_distance_dependent_dielectric yes fps_dielectric 4.0 fps_vdw_fp_scale 1 fps_es_fp_scale 1 fps_hb_fp_scale 0 pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix fps.min.output write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
This script produces a *.txt file to which you can plot the footprint using the provided python script: plot_footprint_single_magnitude.py
to run this enter the following into your command line:
python plot_footprint_single_magnitude.py fps.min.output_footprint_scored.txt 50 ### NOTE: Here, we want the 50 highest contribution interactions. The order of these arguments matter! ###
The result should look something like this:
VI. Virtual Screening
We will create a virtual-screen.in file in 006.virtual-screen that looks like the following:
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file small_ligand_library.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../001.files/lig_withH_charge_1c87.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../004.grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 1c87_virtualscreen write_orientations no num_scored_conformers 1 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
To submit the job for docking, create submit.sh file.
vi submit.sh
Write down the following script in submit.sh file.
#! /bin/bash #PBS -l walltime=48:00:00 #PBS -l nodes=1:ppn=24 #PBS -q long #PBS -N 1c87_rigid #PBS -V cd $PBS_O_WORKDIR dock6 -i rigid.in -o 1c87_rigid_docking.out
After creating this script, submit the job.
qsub submit.sh
We can view whether the job runs or does not run (if there is an error):
qstat -u username
Large Library Virtual Screening (with MPI)
Using 24 processors for two days will not be sufficient to dock all ~25,000 compounds. We can utilize more processors using the Message Passing Interface (MPI) protocol. (This changes the contents of the submission script a little bit)
#! /bin/bash #PBS -l walltime=48:00:00 #PBS -l nodes=4:ppn=28 #PBS -q long #PBS -N 1c87_rigid #PBS -V cd $PBS_O_WORKDIR mpirun -np 112 dock6.mpi -i rigid.in -o 1c87_rigid_docking.out
make sure you are requesting the same number of processors on both cwulf and the MPI command (the -np flag). With these resources, we should finish all 25,000 compounds within one day
Cartesian Minimization of Docked Molecules
Here, cartesian just means we will be using an explicit receptor environment as opposed to the energy grid calculated earlier. These calculations should be calculated in a different sub-directory so first we can create a directory called 007.cartesian-minimization, and create a new min.in file inside that directory.
mkdir 008.cartesian-minimization cd !$ touch min.in
Now we can call dock and answer the questions like before:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file 1c87_virtualscreen_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary yes continuous_score_secondary no cont_score_rec_filename ../001.files/rec_withH_1c87.mol2 cont_score_att_exp 6 cont_score_rep_exp 12 cont_score_rep_rad_scale 1 cont_score_use_dist_dep_dielectric yes cont_score_dielectric 4.0 cont_score_vdw_scale 1 cont_score_es_scale 1 footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 1c87.virtualscreen1.minimized write_orientations no num_scored_conformers 1 rank_ligands no
Since this is another job which needs lots of resources, we submit this to the computing nodes with the submission script. First, create submit.sh.
vi submit.sh
Now, copy the following into the file:
#! /bin/bash #PBS -l walltime=48:00:00 #PBS -l nodes=4:ppn=28 #PBS -q long #PBS -N 1c87_min #PBS -V cd $PBS_O_WORKDIR mpirun -np 112 dock6.mpi -i min.in -o 1c87_vir-screen_cartesian-min.out
Rescoring Docked Molecules
Now, we are going to rescore virtual screen using foorprint similarity, pharmacophore score, tantimoto score and hungarian and the volume overlap score.
touch descriptor.in ../../../zzz.programs/dock6/bin/dock6 -i descriptor.in
Here is the descriptor.in script:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file /gpfs/projects/AMS536/2018/seung/006.virtual-screen.mpi/1c87.flex_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary no footprint_similarity_score_secondary no pharmacophore_score_primary no pharmacophore_score_secondary no descriptor_score_primary yes descriptor_score_secondary no descriptor_use_grid_score no descriptor_use_multigrid_score no descriptor_use_continuous_score no descriptor_use_footprint_similarity yes descriptor_use_pharmacophore_score yes descriptor_use_tanimoto yes descriptor_use_hungarian yes descriptor_use_volume_overlap yes descriptor_fps_score_use_footprint_reference_mol2 yes descriptor_fps_score_footprint_reference_mol2_filename /gpfs/projects/AMS536/2018/seung/005.dock/1c87.min_scored.mol2 descriptor_fps_score_foot_compare_type Euclidean descriptor_fps_score_normalize_foot no descriptor_fps_score_foot_comp_all_residue yes descriptor_fps_score_receptor_filename /gpfs/projects/AMS536/2018/seung/001.files/rec_withH_1c87.mol2 descriptor_fps_score_vdw_att_exp 6 descriptor_fps_score_vdw_rep_exp 12 descriptor_fps_score_vdw_rep_rad_scale 1 descriptor_fps_score_use_distance_dependent_dielectric yes descriptor_fps_score_dielectric 4.0 descriptor_fps_score_vdw_fp_scale 1 descriptor_fps_score_es_fp_scale 1 descriptor_fps_score_hb_fp_scale 0 descriptor_fms_score_use_ref_mol2 yes descriptor_fms_score_ref_mol2_filename /gpfs/projects/AMS536/2018/seung/005.dock/1c87.min_scored.mol2 descriptor_fms_score_write_reference_pharmacophore_mol2 no descriptor_fms_score_write_reference_pharmacophore_txt no descriptor_fms_score_write_candidate_pharmacophore no descriptor_fms_score_write_matched_pharmacophore no descriptor_fms_score_compare_type overlap descriptor_fms_score_full_match yes descriptor_fms_score_match_rate_weight 5.0 descriptor_fms_score_match_dist_cutoff 1.0 descriptor_fms_score_match_proj_cutoff 0.7071 descriptor_fms_score_max_score 20 descriptor_fingerprint_ref_filename /gpfs/projects/AMS536/2018/seung/005.dock/1c87.min_scored.mol2 descriptor_hms_score_ref_filename /gpfs/projects/AMS536/2018/seung/005.dock/1c87.min_scored.mol2 descriptor_hms_score_matching_coeff -5 descriptor_hms_score_rmsd_coeff 1 descriptor_volume_score_reference_mol2_filename /gpfs/projects/AMS536/2018/seung/005.dock/1c87.min_scored.mol2 descriptor_volume_score_overlap_compute_method analytical descriptor_weight_fps_score 1 descriptor_weight_pharmacophore_score 1 descriptor_weight_fingerprint_tanimoto -1 descriptor_weight_hms_score 1 descriptor_weight_volume_overlap_score -1 gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex _drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn chem_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/chem.defn pharmacophore_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/ph4.defn ligand_outfile_prefix descriptor.out write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
Now, copy the following into the file:
#! /bin/bash #PBS -l walltime=48:00:00 #PBS -l nodes=1:ppn=24 #PBS -q long #PBS -N 1c87_rescored #PBS -V cd $PBS_O_WORKDIR dock6 -i descriptor.in -o rescored.out
We are going to submit the job again.
qsub submit.sh.