2019 Denovo design tutorial 2 with PDB 2P16
Contents
I. de-novo design with PDB 2P16 (Focused)
This tutorial assumes that you have finished the Virtual Screening of this system (2P16). We are going to used some of the files generated by the VS toturial.
Fragment Libraries
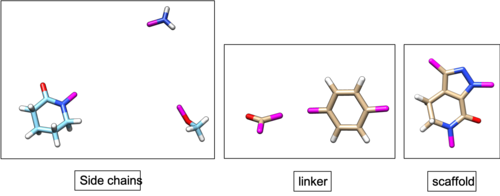
A focused fragment library will be used in this tutorial, in order to attempt building the same ligand. A focused fragment library can be generated using the same ligand.
Create a new directory for the fragment library.
mkdir fraglib
Inside the fraglib directory create a new input file for fragment generation.
touch fraglib.in
Generate the fragments by calling the input file through DOCK6
dock6 -i fraglib.in
Answer the prompted questions interactively using the following lines.
conformer_search_type flex write_fragment_libraries yes fragment_library_prefix fraglib fragment_library_freq_cutoff 1 fragment_library_sort_method freq fragment_library_trans_origin no use_internal_energy yes internal_energy_rep_exp 9 internal_energy_cutoff 100.0 ligand_atom_file (your path for VS docking)/001.files/2p16_wh_lig.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand yes automated_matching yes receptor_site_file (your path for VS docking)/002.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix output write_orientations no num_scored_conformers 1 rank_ligands no
Once fragment generation is completed, following files will be generated. (fraglib_linker.mol2, fraglib_rigid.mol2, fraglib_scaffold.mol2, fraglib_sidechain.mol2, fraglib_torenv.dat) You can use the following commands to check the number of fragments in each file.
grep -wc MOLECULE *.mol2
You can also open the mol2 files using chimera and check if the fragments match with the ligand used for fragment generation.
Focused Denovo Growth
The generated fragments will be used to perform the de novo dock. Here the fragments will added together restricted by different properties like maximum layers, formal charge and molecular weight. Before each connection, DOCK will check with the torsion environment file created in frgment generation to see if that particular connection is seen before. If not the connection won't be made.
Create a new directory for focused library denovo dock.
mkdir focus cd focus
Create a new input file.
touch focus.in
Run the input file through dock.
dock6 -i focus.in
Answer the prompted question interactively using the following lines.
conformer_search_type denovo dn_fraglib_scaffold_file ../fraglib/fraglib_scaffold.mol2 dn_fraglib_linker_file ../fraglib/fraglib_linker.mol2 dn_fraglib_sidechain_file ../fraglib/fraglib_sidechain.mol2 dn_user_specified_anchor no dn_use_torenv_table yes dn_torenv_table ../fraglib/fraglib_torenv.dat dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 1.0 dn_pruning_clustering_cutoff 100.0 dn_constraint_mol_wt 550.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 1 dn_max_grow_layers 9 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_write_checkpoints no dn_write_prune_dump no dn_write_orients no dn_write_growth_trees no dn_output_prefix focus_output use_internal_energy yes internal_energy_rep_exp 9 internal_energy_cutoff 100.0 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file (your path for VS docking)/002.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix (your path for VS docking)/003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl
Focused Denovo Rescore
Create a new directory
mkdir focuseddenovorescore
Copy your reference .mol2 file into this directory
Create an input file
touch rescore.in
Complete the input file using the automated questions from dock
dock6 -i rescore.in
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file output.denovo_build.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary no footprint_similarity_score_secondary no pharmacophore_score_primary no pharmacophore_score_secondary no descriptor_score_primary yes descriptor_score_secondary no descriptor_use_grid_score no descriptor_use_multigrid_score no descriptor_use_continuous_score no descriptor_use_footprint_similarity yes descriptor_use_pharmacophore_score yes descriptor_use_tanimoto yes descriptor_use_hungarian yes descriptor_use_volume_overlap yes descriptor_fps_score_use_footprint_reference_mol2 yes descriptor_fps_score_footprint_reference_mol2_filename candidate_mol_gen_frag.mol2 descriptor_fps_score_foot_compare_type Euclidean descriptor_fps_score_normalize_foot no descriptor_fps_score_foot_comp_all_residue yes descriptor_fps_score_receptor_filename ../../dock/2p16.rec.withH.charged.mol2 descriptor_fps_score_vdw_att_exp 6 descriptor_fps_score_vdw_rep_exp 12 descriptor_fps_score_vdw_rep_rad_scale 1 descriptor_fps_score_use_distance_dependent_dielectric yes descriptor_fps_score_dielectric 4.0 descriptor_fps_score_vdw_fp_scale 1 descriptor_fps_score_es_fp_scale 1 descriptor_fps_score_hb_fp_scale 0 descriptor_fms_score_use_ref_mol2 yes descriptor_fms_score_ref_mol2_filename candidate_mol_gen_frag.mol2 descriptor_fms_score_write_reference_pharmacophore_mol2 no descriptor_fms_score_write_reference_pharmacophore_txt no descriptor_fms_score_write_candidate_pharmacophore no descriptor_fms_score_write_matched_pharmacophore no descriptor_fms_score_compare_type overlap descriptor_fms_score_full_match yes descriptor_fms_score_match_rate_weight 5.0 descriptor_fms_score_match_dist_cutoff 1.0 descriptor_fms_score_match_proj_cutoff 0.7071 descriptor_fms_score_max_score 20 descriptor_fingerprint_ref_filename candidate_mol_gen_frag.mol2 descriptor_hms_score_ref_filename candidate_mol_gen_frag.mol2 descriptor_hms_score_matching_coeff -5 descriptor_hms_score_rmsd_coeff 1 descriptor_volume_score_reference_mol2_filename candidate_mol_gen_frag.mol2 descriptor_volume_score_overlap_compute_method analytical descriptor_weight_fps_score 1 descriptor_weight_pharmacophore_score 1 descriptor_weight_fingerprint_tanimoto -1 descriptor_weight_hms_score 1 descriptor_weight_volume_overlap_score -1 gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file ../../dock/box/vdw_AMBER_parm99.defn flex_defn_file ../../dock/dock/flex.defn flex_drive_file ../../dock/dock/flex_drive.tbl chem_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/chem.defn pharmacophore_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/ph4.defn ligand_outfile_prefix descriptor.output write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
You will now have a file in your directory called descriptor.output_scored.mol2 which can be loaded into Chimera using ViewDock with your original ligand .mol2 file.
II.de-novo growth with PDB 2P16 (Generic)
Running de novo growth
In this section, a de novo growth will be done using the grids generated above and a provided fragment library where the fragments are generated from random drug like ligands provided in the dock6 distribution in the parameters directory.
Go one directory back from multigrid and create a new directory, denovo. Copy the neccesary fragment library files needed into this directory. (scaffold,linker,sidechain,torsion env ..etc) Then create an input file for denovo growth using the lines below.
conformer_search_type denovo dn_fraglib_scaffold_file fraglib_scaffold.mol2 dn_fraglib_linker_file fraglib_linker.mol2 dn_fraglib_sidechain_file fraglib_sidechain.mol2 dn_user_specified_anchor no dn_use_torenv_table yes dn_torenv_table fraglib_torenv.dat dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 1.0 dn_pruning_clustering_cutoff 100.0 dn_constraint_mol_wt 550.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 1 dn_max_grow_layers 4 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_write_checkpoints yes dn_write_prune_dump no dn_write_orients no dn_write_growth_trees no dn_output_prefix output use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../../../dock/2.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary no footprint_similarity_score_secondary no pharmacophore_score_primary no pharmacophore_score_secondary no descriptor_score_primary yes descriptor_score_secondary no descriptor_use_grid_score no descriptor_use_multigrid_score yes descriptor_use_pharmacophore_score no descriptor_use_tanimoto no descriptor_use_hungarian no descriptor_use_volume_overlap no descriptor_multigrid_score_rep_rad_scale 1.0 descriptor_multigrid_score_vdw_scale 1.0 descriptor_multigrid_score_es_scale 1.0 descriptor_multigrid_score_number_of_grids 26 descriptor_multigrid_score_grid_prefix0 ../multigrid/2p16.resid_014 descriptor_multigrid_score_grid_prefix1 ../multigrid/2p16.resid_016 descriptor_multigrid_score_grid_prefix2 ../multigrid/2p16.resid_019 descriptor_multigrid_score_grid_prefix3 ../multigrid/2p16.resid_020 descriptor_multigrid_score_grid_prefix4 ../multigrid/2p16.resid_036 descriptor_multigrid_score_grid_prefix5 ../multigrid/2p16.resid_037 descriptor_multigrid_score_grid_prefix6 ../multigrid/2p16.resid_038 descriptor_multigrid_score_grid_prefix7 ../multigrid/2p16.resid_040 descriptor_multigrid_score_grid_prefix8 ../multigrid/2p16.resid_051 descriptor_multigrid_score_grid_prefix9 ../multigrid/2p16.resid_057 descriptor_multigrid_score_grid_prefix10 ../multigrid/2p16.resid_058 descriptor_multigrid_score_grid_prefix11 ../multigrid/2p16.resid_060 descriptor_multigrid_score_grid_prefix12 ../multigrid/2p16.resid_072 descriptor_multigrid_score_grid_prefix13 ../multigrid/2p16.resid_074 descriptor_multigrid_score_grid_prefix14 ../multigrid/2p16.resid_075 descriptor_multigrid_score_grid_prefix15 ../multigrid/2p16.resid_076 descriptor_multigrid_score_grid_prefix16 ../multigrid/2p16.resid_078 descriptor_multigrid_score_grid_prefix17 ../multigrid/2p16.resid_093 descriptor_multigrid_score_grid_prefix18 ../multigrid/2p16.resid_104 descriptor_multigrid_score_grid_prefix19 ../multigrid/2p16.resid_106 descriptor_multigrid_score_grid_prefix20 ../multigrid/2p16.resid_115 descriptor_multigrid_score_grid_prefix21 ../multigrid/2p16.resid_116 descriptor_multigrid_score_grid_prefix22 ../multigrid/2p16.resid_117 descriptor_multigrid_score_grid_prefix23 ../multigrid/2p16.resid_126 descriptor_multigrid_score_grid_prefix24 ../multigrid/2p16.resid_128 descriptor_multigrid_score_grid_prefix25 ../multigrid/2p16.resid_remaining descriptor_multigrid_score_fp_ref_mol no descriptor_multigrid_score_fp_ref_text yes descriptor_multigrid_score_footprint_text ../2p16.reference.txt descriptor_multigrid_score_foot_compare_type Euclidean descriptor_multigrid_score_normalize_foot no descriptor_multigrid_score_vdw_euc_scale 1.0 descriptor_multigrid_score_es_euc_scale 1.0 descriptor_weight_multigrid_score 1 gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl
Use the following script (denovo.sh) to submit the job to the cluster.
#!/bin/bash #PBS -l walltime=48:00:00 #PBS -l nodes=1:ppn=28 #PBS -q long #PBS -N 2nnq_denovo #PBS -V cd $PBS_O_WORKDIR dock6 -i denovo.in -o 2nnq_denovo.out
Use the following command to submit the job.
qsub denovo.sh