2021 AMBER tutorial 3 with PDBID 1S19
In this tutorial, we will be modeling ligand binding to our receptor using AMBER 16, a molecular dynamics simulation software package created in part by our very own Carlos Simmerling.
Contents
Initial Structures
We will be saving all of our initial structures in a directory called 01.structure.
Protein
For the initial protein structure, we will be using the PDB we downloaded from the Protein Data Bank Website (see here). Load this PDB into chimera and delete the ligand and any nonstandard residues (ex. waters). Save this as a PDB file, 1s19_fresh.pdb, and transfer it to your 01.structure folder.
Ligand
For the ligand, we will again load the 1s19 pdb file from the Protein Data Bank in Chimera. We will delete everything except for the ligand. For this example, the ligand is under the name MC9. To delete everything:
Select -> Residue -> MC9 Select -> Invert (all models) Actions -> Atoms/Bonds -> Delete
With the ligand isolated, we will add hydrogens and charge. To do this:
Tools -> Structure Editing -> Add H Tools -> Structure Editing -> Add Charge -> (have Amber ff14SB and AM1-BCC selected) -> Ok
Save this as a mol2 file in Chimera and save under the name 1s19_ligand_dockprep.mol2.
NOTE It is VERY important to make sure that Chimera adds hydrogens correctly. For this particular ligand the net charge was zero and Chimera was able to model the hydrogens correctly. Many times however, Chimera will add extra hydrogens. If it does, just select the extra hydrogen and go to:
Actions -> Atoms/Bonds -> Delete
Your ligand will then be protonated and charged correctly.
Generating Parameters for the Simulation
Create a new directory called 02.parameters
In order to utilize Amber for molecular dynamics, parameters for the bio molecules will be needed. Luckily, there have been years of parameter development so parameters for the protein do not have to worried about. However, the small ligand does not have parameters in the standard protein force field. Consequently, we will need to generate a fcmod file specific for the ligand.
To do this, we are going to run the following command:
antechamber -i ./../01.structure/1s19_ligand_dockprep.mol2 -fi mol2 -o1s19_ligand_antechamber.mol2 -fo mol2 -at gaff2 -c bcc -rn LIG -nc 0
Gaff2 stands for General Amber Force Field 2, which allows us to generate the parameters for the ligand. The flag at the end -nc stands for net charge. In our case the net charge is zero so we put a zero there, but change it accordingly for your ligand.
Next, we are going to check these parameters and generate a frcmod file with the following command:
parmchk2 -i 1s19_ligand_antechamber.mol2 -f mol2 -o 1s19_ligand.am1bcc.frcmod
This command will generate our 1s19_ligand.am1bcc.frcmod file. It is important to keep checking your output files to ensure that everything looks okay. Once checked, we can move onto the next step.
Building the System with TLeap
Create a new directory called 03.leap and move into it.
Up until now, we have separate models for our protein and our ligand. In order to simulate them as a single system, we have to run tleap. TLeap will generate parameter (parm7) and restart (coordinate - rst7) files. To do this, create the file leap.in and copy in the following script.
#!/usr/bin/sh
###load protein force field
source leaprc.protein.ff14SB
###load GAFF force field (for our ligand)
source leaprc.gaff
###load TIP3P (water) force field
source leaprc.water.tip3p
###load ions frcmod for the tip3p model
loadamberparams frcmod.ionsjc_tip3p
###needed so we can use igb=8 model
set default PBradii mbondi3
###load protein pdb file
rec=loadpdb ./../001.structure/1s19_fresh.pdb
###load ligand frcmod/mol2
loadamberparams ./../002.1.parameters/1s19_ligand.am1bcc.frcmod
lig=loadmol2 ./../002.1.parameters/1s19_ligand_antechamber.mol2
###create gase-phase complex
gascomplex= combine {rec lig}
###write gas-phase pdb
savepdb gascomplex 1s19.gas.complex.pdb
###write gase-phase toplogy and coord files for MMGBSA calc
saveamberparm gascomplex 1s19.complex.parm7 1s19.gas.complex.rst7
saveamberparm rec 1s19.gas.receptor.parm7 1s19.gas.receptor.rst7
saveamberparm lig 1s19.gas.ligand.parm7 1s19.gas.ligand.rst7
###create solvated complex (albeit redundant)
solvcomplex= combine {rec lig}
###solvate the system
solvateoct solvcomplex TIP3PBOX 12.0
###Neutralize system
addions solvcomplex Cl- 0
addions solvcomplex Na+ 0
#write solvated pdb file
savepdb solvcomplex 1s19.wet.complex.pdb
###check the system
charge solvcomplex
check solvcomplex
###write solvated toplogy and coordinate file
saveamberparm solvcomplex 1s19.wet.complex.parm7 1s19.wet.complex.rst7
quit
To run this script, type:
tleap -f leap.in
The first section of the script loads the ff14SB, GAFF and TIP3P force fields. In the second part of the script we load our protein, ligand and ligand parameters. The last part of the script creates our parameter and restart files. Most important to us will be the wet complex files. After the files are generated it is very important to check them in Chimera to make sure they look okay. To load parm7 and rst7 files, you have to go to:
Tools -> MD/Ensemble Analysis -> MD Movie --> Choose your parameter and coordinate files to load in
You should get an image that looks like the one below.
Equilibration
Create a new directory called 04.equil and move into there.
Before we can do a simulation of our system, we first have to do minimizations and equilibrations of it first. We have to do this becuase there could be unfavorable bond angles, bonds or steric clashes that need to be resolved. During this process, we will relax the structure by changing restraints, temperature, pressure, etc.
Input Files
There will be NINE steps to the equilibration and all of the input files are copied below. Copy the files into the neme provided immediately above each of the input scripts.
01.min.mdin
Minimize all the hydrogens &cntrl imin=1, ! Minimize the initial structure ntmin=2, ! Use steepest descent Ryota Added maxcyc=5000, ! Maximum number of cycles for minimization ntb=1, ! Constant volume ntp=0, ! No pressure scaling ntf=1, ! Complete force evaluation ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask="!@H=", ! atoms to be restrained restraint_wt=5.0, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format /
02.equil.mdin
MD simulation &cntrl imin=0, ! Perform MD nstlim=50000 ! Number of MD steps ntb=2, ! Constant Pressure ntc=1, ! No SHAKE on bonds between hydrogens dt=0.001, ! Timestep (ps) ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=1, ! Complete force evaluation ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":!@H=", ! atoms to be restrained restraint_wt=5.0, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on /
03.min.mdin
Minimize all the hydrogens &cntrl imin=1, ! Minimize the initial structure maxcyc=1000, ! Maximum number of cycles for minimization ntb=1, ! Constant volume ntp=0, ! No pressure scaling ntf=1, ! Complete force evaluation ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask="!@H=", ! atoms to be restrained restraint_wt=2.0, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format /
04.min.mdin
Minimize all the hydrogens &cntrl imin=1, ! Minimize the initial structure maxcyc=1000, ! Maximum number of cycles for minimization ntb=1, ! Constant volume ntp=0, ! No pressure scaling ntf=1, ! Complete force evaluation ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask="!@H=", ! atoms to be restrained restraint_wt=0.1, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format /
05.min.mdin
Minimize all the hydrogens &cntrl imin=1, ! Minimize the initial structure maxcyc=1000, ! Maximum number of cycles for minimization ntb=1, ! Constant volume ntp=0, ! No pressure scaling ntf=1, ! Complete force evaluation ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask="!@H=", ! atoms to be restrained restraint_wt=0.05, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format /
06.equil.mdin
MD simulation &cntrl imin=0, ! Perform MD nstlim=50000 ! Number of MD steps ntb=2, ! Constant Pressure ntc=1, ! No SHAKE on bonds between hydrogens dt=0.001, ! Timestep (ps) ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=1, ! Complete force evaluation ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask="!@H=", ! atoms to be restrained restraint_wt=1.0, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on /
07.equil.mdin
MD simulation &cntrl imin=0, ! Perform MD nstlim=50000 ! Number of MD steps ntx=5, ! Positions and velocities read formatted irest=1, ! Restart calculation ntc=1, ! No SHAKE on for bonds with hydrogen dt=0.001, ! Timestep (ps) ntb=2, ! Constant Pressure ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=1, ! Complete force evaluation ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask="!@H=", ! atoms to be restrained restraint_wt=0.5, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on /
08.equil.mdin *Make sure to change the value at restraintmask to the number of residues in your protein including your ligand*
MD simulation &cntrl imin=0, ! Perform MD nstlim=50000 ! Number of MD steps ntx=5, ! Positions and velocities read formatted irest=1, ! Restart calculation ntc=1, ! No SHAKE on for bonds with hydrogen dt=0.001, ! Timestep (ps) ntb=2, ! Constant Pressure ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=1, ! Complete force evaluation ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-433@CA,C,N", ! atoms to be restrained restraint_wt=0.1, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on /
09.equil.mdin *Make sure to change the value at restraintmask to the number of residues in your protein including your ligand*
MD simulation &cntrl imin=0, ! Perform MD nstlim=50000 ! Number of MD steps ntx=5, ! Positions and velocities read formatted irest=1, ! Restart calculation ntc=1, ! No SHAKE on for bonds with hydrogen dt=0.001, ! Timestep (ps) ntb=2, ! Constant Pressure ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=1, ! Complete force evaluation ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-433@CA,C,N", ! atoms to be restrained restraint_wt=0.1, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on /
Run Script
To run all nine steps of the equilibration, we will create a run script with MPI. The run script is copied below with the name of the scrit immediately above.
mdequilibration.sh
#!/bin/sh
#SBATCH --job-name=1s19_equilibration
#SBATCH --ntasks-per-node=40
#SBATCH --nodes=2
#SBATCH --time=8:00:00
#SBATCH -p long-40core
cd $SLURM_SUBMIT_DIR
echo "started Equilibration on 'date' "
do_parallel="mpirun pmemd.MPI"
parm7="./../03.leap/1s19.wet.complex.parm7"
coords="./../03.leap/1s19.wet.complex"
MDINPUTS=( 01.min 02.equil 03.min 04.min 05.min 06.equil 07.equil 08.equil 09.equil)
for input in ${MDINPUTS[@]}; do
$do_parallel -O -i ${input}.mdin -o ${input}.mdout -p $parm7 -c ${coords}.rst7 -ref ${coords}.rst7 -x ${input}.trj -inf ${input}.info -r
${input}.rst7 coords=$input
done
echo " Finished equilibration on 'date' "
To run this script:
sbatch mdequilibration.sh
Important Notes
We ran into a few problems using this run script sometimes but were easily solved. If:
1) Your equilibration gets stuck at a certain step (ex. cannot get past step 6)
Run with pmemd instead of MPI. To change this, delete the do_parallel=mpirun pmemd.MPI. In the for loop, switch out $do_parallel with pmemd.
2) The equilibration will not start, saying that the correct commands are not found.
Try deleting the line cd $SLURM_SUBMIT_DIR
Production
Create a new directory called 05.prod and change into this directory.
When your equilibration is done running, you should double check that the strucutres look okay after the 09.equil step. Repeat the process as with the examination of the 1s19.wet.complex files in Chimera during the tleap step. If everything looks okay, then you are good to go to continue ot the production runs.
Input File
Below is the input file for 10.prod.mdin
For the restraint mask, take out your ligand (so only include protein)
(To do this as an unrestrained simulation, just delete the restraint mask and weight as a whole)
MD simulations &cntrl imin=0, ! Perform MD nstlim=5000000, ! Number of MD steps ntx=5, ! Positions and velocities read formatted irest=1, ! Restart calculation ntc=2, ! SHAKE on for bonds with hydrogen dt=0.002, ! Timestep (ps) ntb=2, ! Constant Pressure ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=2, ! No force evaluation for bonds with hydrogen ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 2500, ! Write to trajectory file every ntwx steps ntpr= 2500, ! Print to mdout every ntpr steps ntwr= 5000000, ! Write a restart file every ntwr steps cut=8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-432@CA,C,N", ! atoms to be restrained restraint_wt=0.1, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on /
This step is very similar to the 09.equli.mdin input file. The only things that changed was nstlim, ntc, dt, ntwx, ntpr and ntwr lines.
Run Script
Below is the run script that can be copied into a file called mdproduction.sh
#!/bin/sh
#SBATCH --job-name=1s19_prod
#SBATCH --ntasks-per-node=24
#SBATCH --nodes=2
#SBATCH --time=7-00:00:00
#SBATCH -p extended-24core
cd $SLURM_SUBMIT_DIR
echo "started Equilibration on 'date' "
parm7="../03.leap/1s19.wet.complex.parm7"
coords="../04.equil/09.equil"
MDINPUTS=( 10.prod)
for input in ${MDINPUTS[@]}; do
pmemd -O -i ${input}.mdin -o ${input}.mdout -p ${parm7} -c ${coords}.rst7 -ref ${coords}.rst7 -x ${input}.trj -inf ${input}.info -r
${input}.rst7
coords=$input
done
echo "Finished Equilibration on `date` "
To run this script:
sbatch mdproduction.sh
Analysis
Now that we have created the MD trajectory, we want to run an analysis of how the ligand-receptor system has changed and what interactions are critical for ligand binding. We will do this by measuring RMSD, H-bonds, and MM-GBSA.
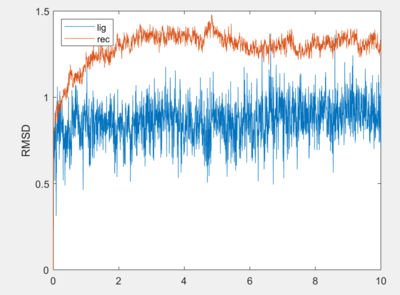
RMSD
Root-mean-square deviations depict how far the ligand and receptor have moved from their starting position. We will begin by creating the following input files:
touch cpptraj_rmsd_lig.in, cpptraj_rmsd_rec.in, cpptraj_strip_wat.in
First we will strip the trajectory of all waters and ions.
vim cpptraj_strip_wat.in
#!/usr/bin/sh parm ../003.tleap/1s19.wet.complex.parm7 #read in trajectory trajin ../004.productionrun/10.prod.trj #read in reference reference ../003.tleap/1s19.wet.complex.rst7 #compute RMSD + align CA to crystal structure rmsd rms1 reference :1-263@CA #strip solvent strip :WAT:Na+:Cl- #create gas-phase trajectory trajout 1S19_stripfit_restrained_gas.trj nobox
cpptraj -i cpptraj_strip_wat.in
Then we will measure the RMSD of the ligand and receptor respectively.
vim cpptraj_rmsd_lig.in
#!/usr/bin/sh #trajin the trajectory trajin 1S19_stripfit_restrained_gas.trj #read in reference reference ../003.tleap/1s19.gas.complex.rst7 #compute RMSD (do not fit internal geometris first, included rigid body motions, convert frames to ns (framenum*.005) rmsd rms1 ":254&!(@H=)" nofit mass out 1S19_lig_restrained_rmsd_nofit.dat time .005 #histogram the nofit rmsd histogram rms1,*,*,.1,* norm out 1S19_lig_restrained_rmsd_nofit_histogram.dat
Make sure to change the number of the ligand in the "rmsd rms1" line to match your particular system To submit the job:
cpptraj -p ../003.tleap/1s19.gas.complex.parm7 -i cpptraj_rmsd_lig.in
vim cpptraj_rmsd_rec.in
#!/usr/bin/sh #trajin the trajectory trajin 1S19_stripfit_restrained_gas.trj #read in reference reference ../003.tleap/1s19.gas.complex.rst7 #compute RMSD (do not fit internal geometries first, included rigid body motions, convert frames to ns (framenum*.005) rmsd rms1 ":1-253&!(@H=)" nofit mass out 1S19_rec_restrained_rmsd_nofit.dat time .005 #histogram the nofit rmsd histogram rms1,*,*,.1,* norm out 1S19_rec_restrained_rmsd_nofit_histogram.dat
Make sure to change the residues of the protein in the "rmsd rms1" line to match your particular system To submit the job:
cpptraj -p ../003.tleap/1s19.gas.complex.parm7 -i cpptraj_rmsd_rec.in
You should get an output that looks like the one below.
Hydrogen Bonds
Here we want to figure out what hydrogen bonds our ligand forms with our receptor. We will begin by creating an input file:
gedit cpptraj_hbond.in
!/usr/bin/sh #read in trajectory trajin ../004.productionrun/10_prod.trj #wrap everything in one periodic cell - could cause problems, may comment out #autoimage if problems later autoimage #compute intra + water-mediated H-bonds hbond hb1 :1-263 out 1S19_calcipotriol_hbond.out avgout 1S19_calcipotriol_hbond_avg.dat solventdonor :WAT solventacceptor :WAT@-O nointramol brid\ geout 1S19_calcipotriol_bridge-water.dat dist 3.0 angle 140
Run this input file using the same cpptraj tool:
cpptraj -p ../002.tleap_build/1S19_wetcomplex.parm7 -i cpptraj_hbond.in
MMGBSA
Molecular Mechanics-Generalized Born Solvent Accessibility is a method to estimate the free energy of binding of our ligand and receptor by evaluating the free energy of unsolvated and solvated ligand and receptor separately then combining them. To measure this binding, we will first create an input file:
gedit mmgbsa.in
mmgbsa 1S19 analysis &general interval=1, netcdf=1, keep_files=0, / &gb igb=8 saltcon=0.0, surften=0.0072, surfoff=0.0, molsurf=0, / &nmode drms=0.001, maxcyc=10000, nminterval=250, nmendframe=2000, nmode_igb=1, /
We need to create a SLURM script to run this input file. Create the following file mmgbsa_1S19_slurm.sh:
#bin/bash
#SBATCH --time=2-00:00:00
#SBATCH --nodes=2
#SBATCH --ntasks=28
#SBATCH --job-name=1S19_MMGBSA
#SBATCH --output=1S19_MMGBSA.log
#SBATCH -p extended-28core
#Define topology files
solv_parm="../002.tleap_build/1S19_wetcomplex.parm7"
complex_parm="../002.tleap_build/1S19_gascomplex.parm7"
receptor_parm="../002.tleap_build/1S19_gasrec.parm7"
lig_parm="../002.tleap_build/1S19_gaslig.parm7"
trajectory="../004.productionrun/10_prod.trj"
MMGBSA.py -O -i mmgbsa.in \
-o 1S19_mmgbsa_results.dat \
-eo 1S19_mmgbsa_perframe.dat \
-sp ${solv_parm} \
-cp ${complex_parm} \
-rp ${receptor_parm} \
-lp ${lig_parm} \
-y ${trajectory}