2025 DOCK GA tutorial 2 with 1XMU
Contents
Introduction
Introduced in DOCK6.10 is the genetic algorithm, a form of de novo drug design that employs molecular evolution (mutations) and an iterative natural selection process. Several fragment-based (requiring a fragment library) mutations, including cross-over, addition, deletion, substitution, and replacement, are used on the provided “parent” molecule(s) to produce a new generation of “off-spring”. Subsequently, the natural selection process uses user-defined variables to exclude “off-spring” with poor scoring (and/or lesser fitness) from being included in the next generation of “parents”.
In this section of the DOCK6.12 tutorial, we will:
- generate a fragment library
- run a genetic algorithm
- perform an analysis using USCF Chimera
Set Up
Before we begin, we should create separate directories to guide and organize our workflow.
mkdir 00X_fragLib mkdir 00X_algorithm
Generating the Library
The genetic algorithm requires a pre-docked fragment library - an ensemble of scaffolds, linkers, and sidechains, used to mutate the ligand during the genetic algorithm. DOCK6.12 allows for the creation of a personalized fragment library via deconstruction of a chosen molecule (here, our chosen ligand).
Creating a Library Input File
Enter your new directory, create an empty input file, and access it with dock6:
cd 00X_fragLib touch 1XMU_fragLib.in dock6 -i 1XMU_fragLib.in
Answer the prompts using the following responses. You can also modify the example code below to match your needs.
Note: Depending on the version of dock6 that you are using, some prompts displayed here may not exist anymore, or you might encounter newer/older prompts not present.
conformer_search_type flex write_fragment_libraries yes fragment_library_prefix 1XMU_fragLib fragment_library_freq_cutoff 1 fragment_library_sort_method freq fragment_library_trans_origin no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../00X_structure/1XMU_Lig_wCH.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules no atom_model all vdw_defn_file /PATH/vdw_AMBER_parm99.defn flex_defn_file /PATH/parameters/flex.defn flex_drive_file /PATH/flex_drive.tbl ligand_outfile_prefix 1XMU_output write_mol_solvation no write_orientations no num_scored_conformers 1 score_threshold 100.0 rank_ligands no
If you encounter any issues, you can always edit the file with vi/vim and then run it using:
dock6 -i 1XMU_fragLib.in -o 1XMU_fragLib.out>&1XMU_fragLib.log&
Note: This will create additional output (1XMU_fragLib.out) and log (1XMU_fragLib.log) files, which may be useful to discern errors in the input.
This process will take a few seconds and output several files:
- 1XMU_fraglib_rigid.mol2
- 1XMU_fraglib_scaffold.mol2
- 1XMU_fraglib_sidechain.mol2
- 1XMU_fraglib_linker.mol2
- 1XMU_fraglib_torenv.dat, the torsion environment file
- 1XMU_output_scored.mol2
Combining the Torsion Environment Tables
Before we continue, it is important that we update our torsion environment table. The python script, combine_torenv.py (available in the bin directory) creates a combination of two torenv.dat files.
python ${DOCK_DIR}/bin/combine_torenv.py 1XMU_fraglib_torenv.dat ${DOCK_DIR}/parameters/fraglib_torenv.dat
This will give us a “master” fragment library file (referred to as unique_full_sorted_fraglib.dat).
Running the Genetic Algorithm
Creating a Genetic Input File
Enter your next directory, create an empty input file and access it with dock6:
cd ../00X_algorithm touch 1XMU_geneticAlgo.in dock6 -i 1XMU_geneticAlgo.in
Answer the prompts using the following responses:
Note: Depending on the version of dock6 that you are using, some prompts displayed here may not exist anymore, or you might encounter newer/older prompts not present.
conformer_search_type genetic ga_molecule_file ../../00X_structure/1XMU_Lig_wCH.mol2 ga_utilities no ga_fraglib_scaffold_file /PATH/fraglib_ga_scaffold.mol2 ga_fraglib_linker_file /PATH/fraglib_linker.mol2 ga_fraglib_sidechain_file /PATH/fraglib_sidechain.mol2 ga_torenv_table ../00X_fragLib/unique_full_sorted_fraglib.dat ga_max_generations 100 ga_xover_on yes ga_xover_sampling_method_rand yes ga_xover_max 150 ga_bond_tolerance 0.5 ga_angle_cutoff 0.14 ga_check_overlap no ga_mutate_addition yes ga_mutate_deletion yes ga_mutate_substitution yes ga_mutate_replacement yes ga_mutate_parents yes ga_pmut_rate 0.3 ga_omut_rate 0.7 ga_max_mut_cycles 5 ga_mut_sampling_method rand ga_num_random_picks 15 ga_max_root_size 5 ga_energy_cutoff 100 ga_heur_unmatched_num 1 ga_heur_matched_rmsd 0.5 ga_constraint_mol_wt 500 ga_constraint_rot_bon 10 ga_constraint_H_accept 10 ga_constraint_H_don 5 ga_constraint_formal_charge 2 ga_ensemble_size 100 ga_selection_method elitism ga_elitism_combined yes ga_elitism_option max ga_max_num_gen_with_no_crossover 25 ga_name_identifier ga ga_output_prefix ga_output use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_lig_efficiency no grid_score_grid_prefix ../00X_gridbox/grid minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_final_min no simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /PATH/vdw_AMBER_parm99.defn flex_defn_file /PATH/flex.defn flex_drive_file /PATH/flex_drive.tbl
Accessing a Slurm Queue
This process, depending on your responses, may use significant computational resources. It would be beneficial to access a partition via. slurm queue (if you are utilizing an external cluster).
If submitted to your head node, you can use the kill command to cancel it.
You can modify the example code below to match your needs and system specifications:
#!/bin/bash # #SBATCH --job-name=1XMU_genetic #SBATCH --ntasks-per-node=1 #SBATCH --nodes=1 #SBATCH --time=4:00:00 #SBATCH -p short-96core dock6 -i 1XMU_geneticAlgo.in -o 1XMU_geneticAlgo.out
Submit your job to the queue by typing:
sbatch job.slurm
Note: You can modify the job.slurm script to notify you of failures. Alternatively, you can utilize the grep command to track the generational progression of the genetic algorithm.
This process will take some time and output a large number of files, including:
- 1XMU_geneticAlgo.out
- ga_output.restart0000.mol2, the initial parent ensemble
- ga_output.restartXXXX.mol2, a set of molecules from each generation
Analysis / Conclusion
Downloading the Necessary Files
From this point, you can download (onto your local computer, using the scp command) any of the ga_output.restart files individually, or use the cat command and download the resulting file.
On your local computer, type:
scp netID@network.instuition.edu:/PATH/FILE/ /LOCATION/
Note: For very large molecular evolution processes, it may be useful to create a tarball. You can utilize additional compression tools, if desired.
While in the main directory, type:
tar -cvf 00X_algorithm algorithm.tar
On you local computer, type:
scp netID@network.instuition.edu:/PATH/algorithm.tar /LOCATION/ tar -xvf algorithm.tar
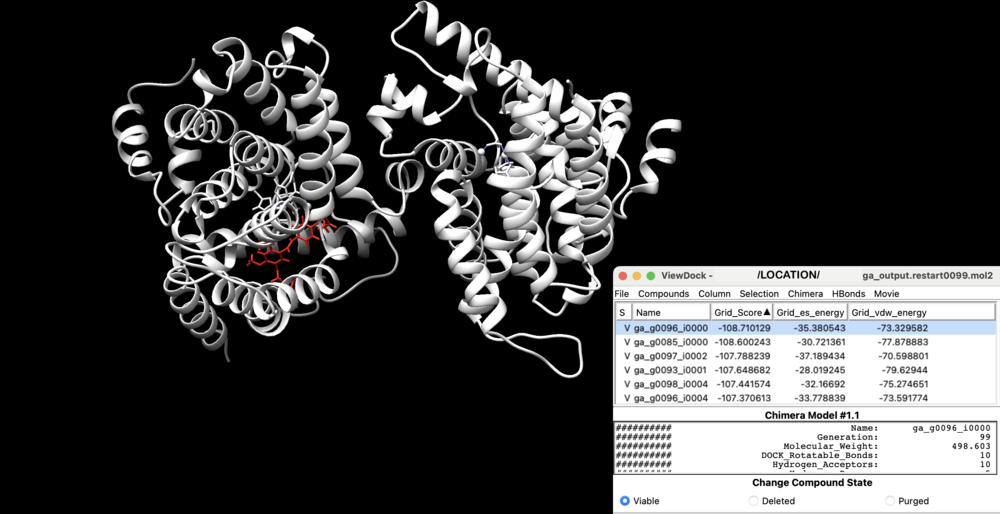
Analyzing the Genetic Algorithm Models
Analysis can then be performed using the UCSF Chimera visualization program. Navigate to the receptor.mol2 file’s location using : Browse (alternatively, File > Open)
Then, access the chosen generation (or concatenated) file using : ViewDock (found under Tools > Surface/Binding Analysis > ViewDock).
You may need to designate a file type for this step (in this case, choose the DOCK 4, 5, or 6 option).
You can now:
- view each ligand molecule’s model within the generation
- add additional columns, to show grid scoring and molecule properties (Column > Show)
- save images, mol2, or pdb files (File > Save Image, PDB, Mol2), as needed