Difference between revisions of "2023 Denovo tutorial 2 with PDBID 3WZE"
Stonybrook (talk | contribs) (→Checking the Results) |
Stonybrook (talk | contribs) (→Rescoring the Outputs) |
||
| (44 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
This tutorial is a continuation of the virtual screening tutorial. In this tutorial, we'll continue to work with the receptor and ligand in PDB 3WZE, and we'll attempt to generate new ligands for the receptor using three kinds of ''de novo'' design: ''de novo'' refinement, focused ''de novo'' design, and generic ''de novo'' design. | This tutorial is a continuation of the virtual screening tutorial. In this tutorial, we'll continue to work with the receptor and ligand in PDB 3WZE, and we'll attempt to generate new ligands for the receptor using three kinds of ''de novo'' design: ''de novo'' refinement, focused ''de novo'' design, and generic ''de novo'' design. | ||
| − | ''De novo'' can be directly translated as "of new", but a more deft translation might be "from the beginning" or "from scratch". This method of ligand generation involves procedurally generating | + | ''De novo'' can be directly translated as "of new", but a more deft translation might be "from the beginning" or "from scratch". This method of ligand generation involves procedurally generating a ligand using algorithms within programs like DOCK, and is typically used to build entirely new ligands for proteins by building molecules outwards from an initial anchor one moiety at a time. |
Generic ''de novo'' design best matches the prior description, in which a pre-selected or random anchor is positioned within the active site of the receptor, and then built outwards in a number of layers occupied by various sampled moieties. Focused ''de novo'' design is much like generic ''de novo'' design, except that the pool of sampled moieties is curtailed to suit the needs of the researcher. Finally, ''de novo'' refinement is when one begins with an already discovered ligand, then deletes some of the molecule and replaces it with a dummy atom, effectively using the remainder of the ligand as the anchor for the ''de novo'' design algorithms to modify. | Generic ''de novo'' design best matches the prior description, in which a pre-selected or random anchor is positioned within the active site of the receptor, and then built outwards in a number of layers occupied by various sampled moieties. Focused ''de novo'' design is much like generic ''de novo'' design, except that the pool of sampled moieties is curtailed to suit the needs of the researcher. Finally, ''de novo'' refinement is when one begins with an already discovered ligand, then deletes some of the molecule and replaces it with a dummy atom, effectively using the remainder of the ligand as the anchor for the ''de novo'' design algorithms to modify. | ||
| Line 8: | Line 8: | ||
=='''Directories'''== | =='''Directories'''== | ||
| − | Make a new directory for ''de novo'' | + | Make new directories for ''de novo'' design: |
| − | + | mkdir 009_denovo_generic | |
| + | mkdir 010_denovo_refine | ||
| + | mkdir 011_denovo_focused | ||
| + | |||
| + | ='''Generic ''De Novo'' Design'''= | ||
| + | |||
| + | In this tutorial, we'll use the generic fragment library distributed with DOCK 6.10, and we won't use a user-specified anchor either. Thus, DOCK will build new ligands for our receptor from scratch. | ||
| + | |||
| + | 1. Bring the grid nrg and bmp files for 3WZE's receptor to your current directory. These files are all that you'll need for a generic ''de novo'' design run performed within the 3WZE receptor's active site. | ||
| + | |||
| + | 2. Type "touch 3WZE_generic.in" to make a blank file. | ||
| + | |||
| + | 3. Type "dock6 -i 3WZE_generic.in" to fill out DOCK's question tree and generate an input file. Use the input file below as a guide for how to answer DOCK's questions: | ||
| + | |||
| + | |||
| + | conformer_search_type denovo | ||
| + | dn_fraglib_scaffold_file | ||
| + | /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_scaffold.mol2 | ||
| + | dn_fraglib_linker_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_linker.mol2 | ||
| + | dn_fraglib_sidechain_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_sidechain.mol2 | ||
| + | dn_user_specified_anchor no | ||
| + | dn_torenv_table /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_torenv.dat | ||
| + | dn_name_identifier 3WZE_generic | ||
| + | dn_sampling_method graph | ||
| + | dn_graph_max_picks 30 | ||
| + | dn_graph_breadth 3 | ||
| + | dn_graph_depth 2 | ||
| + | dn_graph_temperature 100.0 | ||
| + | dn_pruning_conformer_score_cutoff 100.0 | ||
| + | dn_pruning_conformer_score_scaling_factor 2.0 | ||
| + | dn_pruning_clustering_cutoff 100.0 | ||
| + | dn_mol_wt_cutoff_type soft | ||
| + | dn_upper_constraint_mol_wt 550.0 | ||
| + | dn_lower_constraint_mol_wt 0.0 | ||
| + | dn_mol_wt_std_dev 35.0 | ||
| + | dn_constraint_rot_bon 15 | ||
| + | dn_constraint_formal_charge 2.0 | ||
| + | dn_heur_unmatched_num 1 | ||
| + | dn_heur_matched_rmsd 2.0 | ||
| + | dn_unique_anchors 1 | ||
| + | dn_max_grow_layers 9 | ||
| + | dn_max_root_size 25 | ||
| + | dn_max_layer_size 25 | ||
| + | dn_max_current_aps 5 | ||
| + | dn_max_scaffolds_per_layer 1 | ||
| + | dn_write_checkpoints yes | ||
| + | dn_write_prune_dump no | ||
| + | dn_write_orients no | ||
| + | dn_write_growth_trees no | ||
| + | dn_output_prefix 3WZE_generic | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | use_database_filter no | ||
| + | orient_ligand yes | ||
| + | automated_matching yes | ||
| + | receptor_site_file 3WZE_spheres.sph | ||
| + | max_orientations 1000 | ||
| + | critical_points no | ||
| + | chemical_matching no | ||
| + | use_ligand_spheres no | ||
| + | bump_filter yes | ||
| + | bump_grid_prefix grid | ||
| + | max_bumps_anchor 2 | ||
| + | max_bumps_growth 2 | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | grid_score_primary yes | ||
| + | grid_score_rep_rad_scale 1 | ||
| + | grid_score_vdw_scale 1 | ||
| + | grid_score_es_scale 1 | ||
| + | grid_score_grid_prefix grid | ||
| + | minimize_ligand yes | ||
| + | minimize_anchor yes | ||
| + | minimize_flexible_growth yes | ||
| + | use_advanced_simplex_parameters no | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_anchor_max_iterations 500 | ||
| + | simplex_grow_max_iterations 250 | ||
| + | simplex_grow_tors_premin_iterations 0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_de_novo.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl | ||
| + | |||
| + | 4. Terminate the run with control c, then type "nano 3WZE_generic.sh". We're going to run this job on the cluster because it will be too computationally costly for the head node. | ||
| + | |||
| + | 5. Copy the following into the file: | ||
| + | |||
| + | #!/bin/bash | ||
| + | #SBATCH --job-name=3WZE_generic | ||
| + | #SBATCH --ntasks-per-node=24 | ||
| + | #SBATCH --nodes=1 | ||
| + | #SBATCH --time=48:00:00 | ||
| + | #SBATCH -p long-24core | ||
| + | dock6 -i 3WZE_generic.in | ||
| + | |||
| + | 6. Type "sbatch 3WZE_generic.sh" to start the ''de novo'' run on the cluster. The job will probably take 1-2 hours to complete. | ||
| + | |||
| + | 7. Move the "3WZE_generic.denovo_build.mol2" file to your local machine, and either open Chimera and use the viewdock command to open it, or open it in ChimeraX and type "viewdock". There should be approximately 700 ligands produced, and the one with the lowest grid score from our run is shown below: | ||
| + | |||
| + | [[File: Sunodillam.png|thumb|center|1000px]] | ||
| + | |||
| + | =='''Rescoring the Outputs'''== | ||
| + | |||
| + | The new ligands generated by this ''de novo''' run have an assocaited grid score to judge their efficacy in binding to the receptor they were designed for, but each ligand can still be energy minimized by rigid docking to arrive at a more accurate estimate of their ability to bind. | ||
| + | |||
| + | 1. Type "touch 3WZE_min.in" | ||
| + | |||
| + | 2. Type "dock6 -i 3WZE_min.in" and answer the question tree using the following input file as a guide: | ||
| + | |||
| + | conformer_search_type rigid | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file 3WZE_generic.denovo_build.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter no | ||
| + | orient_ligand yes | ||
| + | automated_matching yes | ||
| + | receptor_site_file 3WZE_sphere.sph | ||
| + | max_orientations 1000 | ||
| + | critical_points no | ||
| + | chemical_matching no | ||
| + | use_ligand_spheres no | ||
| + | bump_filter yes | ||
| + | bump_grid_prefix grid | ||
| + | max_bumps_anchor 2 | ||
| + | max_bumps_growth 2 | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | grid_score_primary yes | ||
| + | grid_score_rep_rad_scale 1 | ||
| + | grid_score_vdw_scale 1 | ||
| + | grid_score_es_scale 1 | ||
| + | grid_score_grid_prefix grid | ||
| + | minimize_ligand yes | ||
| + | simplex_max_iterations 1000 | ||
| + | simplex_tors_premin_iterations 0 | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_de_novo.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl | ||
| + | ligand_outfile_prefix 3WZE_generic_min | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| + | |||
| + | 3. Bring the "3WZE_generic_min_scored.mol2" file, which contains the minimized molecules to your local machine, and open it with Chimera or ChimeraX as described previously. Shown below is the ligand with the lowest continuous score after energy minmization: | ||
| + | |||
| + | [[File: Haklamabi.png|thumb|center|1000px]] | ||
| + | |||
| + | If you compare this ligand to the best scored ligand from the generic ''de novo'' design, you'll see that this one actually has a few moieties that are different, suggesting that this molecule is truly best suited for the receptor's binding site, and the prior molecule was only scored highest because all 700 ligands hadn't been energy minimized. | ||
='''''De Novo'' Refinement'''= | ='''''De Novo'' Refinement'''= | ||
| Line 15: | Line 185: | ||
=='''Ligand Preparation'''== | =='''Ligand Preparation'''== | ||
| − | 1. Open the final, energy minimized ligand mol2 file which was used for the virtual screen tutorial, and also open the final receptor mol2 file that was used in that screen. Either Chimera or ChimeraX can be used to open the files. As long as no translations or rotations have occurred during the virtual screen process, the ligand should still be in its native orientation within the receptor's active site, as depicted by the original 3WZE pdb file. | + | 1. Open the final, energy minimized ligand mol2 file which was used for the 3WZE virtual screen tutorial, and also open the final receptor mol2 file that was used in that screen. Either Chimera or ChimeraX can be used to open the files. As long as no translations or rotations have occurred during the virtual screen process, the ligand should still be in its native orientation within the receptor's active site, as depicted by the original 3WZE pdb file. |
2. Examine the binding pocket of the receptor, and choose a part of the 3WZE ligand that faces towards the interior of the binding pocket. Parts of the ligand that are innermost to the receptor make for the best parts to delete because they tend to have the most potential interactions with the protein, allowing the various groups tested in ''de novo'' design to have a better chance of interacting with a group on the protein. Choosing a part of the ligand to delete which faces the cytosol or the channel leading to the cytosol will be less likely to yield new ligands that can bind tightly to the interior of the receptor. To help recognize good sites for deletion, it's a good idea to show sidechains and hbonds, which can allow you to see which parts of the ligand are interacting with the protein. | 2. Examine the binding pocket of the receptor, and choose a part of the 3WZE ligand that faces towards the interior of the binding pocket. Parts of the ligand that are innermost to the receptor make for the best parts to delete because they tend to have the most potential interactions with the protein, allowing the various groups tested in ''de novo'' design to have a better chance of interacting with a group on the protein. Choosing a part of the ligand to delete which faces the cytosol or the channel leading to the cytosol will be less likely to yield new ligands that can bind tightly to the interior of the receptor. To help recognize good sites for deletion, it's a good idea to show sidechains and hbonds, which can allow you to see which parts of the ligand are interacting with the protein. | ||
| Line 23: | Line 193: | ||
In this image, one can see the ligand sorafinib, and also the two hbonds that it forms with the nearby glutamic acid residue 71. It also forms an hbond with the backbone of the receptor using its amide oxygen. Based on this, we'll truncate those two amides and the entire aromatic ring closest to the camera. The camera is positioned to look from the side of the receptor where the binding pocket is deepest, so deleting everything closer than those amides will delete the parts of the ligand which are innermost. | In this image, one can see the ligand sorafinib, and also the two hbonds that it forms with the nearby glutamic acid residue 71. It also forms an hbond with the backbone of the receptor using its amide oxygen. Based on this, we'll truncate those two amides and the entire aromatic ring closest to the camera. The camera is positioned to look from the side of the receptor where the binding pocket is deepest, so deleting everything closer than those amides will delete the parts of the ligand which are innermost. | ||
| − | 3. Select and | + | 3. Select and delete the receptor. Now that we've identified which part of the ligand to remove, we don't need the receptor anymore. |
4. Orient the ligand so that the area you wish to delete is easy to see. Hold control down on your keyboard, then click and drag to cover the area. This should select the area. | 4. Orient the ligand so that the area you wish to delete is easy to see. Hold control down on your keyboard, then click and drag to cover the area. This should select the area. | ||
| Line 43: | Line 213: | ||
8. Open the mol2 file in a text editor on desktop, or with a command like "nano" from the command line. | 8. Open the mol2 file in a text editor on desktop, or with a command like "nano" from the command line. | ||
| − | 9. Find N14, and change the atom type to "Du1". Also change its bond type to "Du". | + | 9. Find N14, and change the atom type to "Du1". Also change its bond type to "Du". We're only adding a single dummy atom to the anchor in this tutorial because we're trying to modify only one part of an exissting ligand, but bear in mind that one could add any number of dummy atoms to an input ligand and DOCK will try adding moieties to each one. |
[[File: Dougdenovo5.png|thumb|center|1000px]] | [[File: Dougdenovo5.png|thumb|center|1000px]] | ||
| Line 53: | Line 223: | ||
(Note that this image was taken with ChimeraX) | (Note that this image was taken with ChimeraX) | ||
| − | 11. Now that our ligand is prepared, we can move it to Seawulf where we can perform the actual ''de novo'' refinement. | + | 11. Now that our ligand is prepared, we can move it to Seawulf where we can perform the actual ''de novo'' refinement. For information on how to move a file to Seawulf using the scp command, see the 3WZE virtual screen tutorial. |
=='''Running the Refinement'''== | =='''Running the Refinement'''== | ||
| Line 59: | Line 229: | ||
As with the virtual screen, DOCK can be run with an input file, the text of which will be shown below. However, it's a good idea to make your own input file rather than copying what is written here. That way, you can get a sense of what parameters can be adjusted before a ''de novo'' refinement run. | As with the virtual screen, DOCK can be run with an input file, the text of which will be shown below. However, it's a good idea to make your own input file rather than copying what is written here. That way, you can get a sense of what parameters can be adjusted before a ''de novo'' refinement run. | ||
| − | 1. In the command line in Seawulf, type | + | 1. In the command line in Seawulf, type |
| + | touch de_novo_refine.in | ||
| + | |||
| + | Unlike "nano" or "vi", the "touch" command will allow you to make a blank file. | ||
2. To go through the process of answering DOCK's many questions about your run, and to subsequently generate an input file, type | 2. To go through the process of answering DOCK's many questions about your run, and to subsequently generate an input file, type | ||
| Line 100: | Line 273: | ||
dn_write_orients no | dn_write_orients no | ||
dn_write_growth_trees no | dn_write_growth_trees no | ||
| − | dn_output_prefix | + | dn_output_prefix 3WZE_refine |
use_internal_energy yes | use_internal_energy yes | ||
internal_energy_rep_exp 12 | internal_energy_rep_exp 12 | ||
| Line 133: | Line 306: | ||
flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn | ||
flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl | ||
| + | |||
| + | Notes on some of the parameters: | ||
| + | |||
| + | -'''dn_user_specified_anchor''' can be set to "no" if one wishes to run a ''de novo'' design run in which they do not specify an input anchor with dummy atoms. When set to "no", DOCK 6.10 will use moieties from the fragment libraries as anchors to build out from, allowing for multiple different anchors and orientations to be tried. | ||
| + | |||
| + | -'''dn_sampling_method''' can be "graph", "random", or "exhaustive". "graph" will select moieties which are similar to previously selected moieties that improved the grid score, so graph will attempt to bias future moiety selection in a way that promotes improving the ligand. "random" will cause moieties to be selected at random. "exhaustive" will ensure that every possible moiety will be tried at every possible position. Be careful with "exhaustive" though, because it will increase the computation time required in a way proportional to the fragment library size. | ||
| + | |||
| + | -'''dn_graph_max_picks''' and '''dn_num_random_picks''' (the latter is not shown above) control how many moieties are tried per dummy atom per growth layer. | ||
| + | |||
| + | -'''dn_max_grow_layers''' controls how many moieties outwards DOCK will grow your ligand from each dummy atom on the initial anchor. We've only set it to 1 in this refinement tutorial for simplicity, but it can be set higher values. 8 to 9 layers is common for growing ligands from scratch without a set anchor, but the total number of layers that you may want will depend on the goal of the ''de novo'' design run. | ||
| + | |||
| + | -'''minimize_ligand''', when set to yes, will attempt to energy minimize the ligand after each moiety selection, in much the same way DOCK 6.10 does so in flexible docking. | ||
| + | |||
| + | -'''minimize_anchor''', when set to yes, will try to energy minimize the anchor before attaching moieties, irrespective of whether the anchor is provided by the user or is chosen randomly. For ''de novo'' runs in which the anchor is part of a ligand that binds in a known orientation, it is best to set this parameter to "no". Otherwise, DOCK 6.10 might alter the orientation of your anchor before attempting to grow it with new moieties, and a resulting molecule might not be reflective of how the ligand normally binds. | ||
| + | |||
| + | -Much like the prior parameter, '''simplex_restraint_min''' is useful when running a ''de novo'' design with a user-supplied anchor of known orientation. When set to yes, this parameter essentially applies a stretchy tether to the initial anchor, allowing it to deviate from its starting position somewhat for the sake of energy minimization, but also applies a penalty based on increasing RMSD. | ||
| + | |||
4. Run the ''de novo'' refinenment in Seawulf using the following command: | 4. Run the ''de novo'' refinenment in Seawulf using the following command: | ||
| Line 148: | Line 338: | ||
2. If you're using Chimera, open Chimera and use Tools->Surface/Binding Analysis->ViewDock to open 3WZE_refine.denovo_build.mol2. If you're using ChimeraX, open that file, then type "viewdockx" into the command line. | 2. If you're using Chimera, open Chimera and use Tools->Surface/Binding Analysis->ViewDock to open 3WZE_refine.denovo_build.mol2. If you're using ChimeraX, open that file, then type "viewdockx" into the command line. | ||
| − | [[File: Dougrefine1.png|thumb|center]] | + | [[File: Dougrefine1.png|thumb|center|1000px]] |
| + | Note that this image was taken from ChimeraX. Also, please disregard that I named my ligand "blooble". | ||
| + | |||
| + | 3. Look through the results. Because we set "dn_max_grow_layers" to 1, this means that the dummy atom should only be replaced with a single fragment, and there should be no further fragments appended to the single one added. | ||
| + | |||
| + | For our results, we got 10 new molecules. If we had wanted more molecules, we could have increased the value of "dn_graph_max_picks" from 30 to a higher value. This means that dock would sample more fragments (We only got 10 molecules because 20 of the 30 picked fragments must have somehow been incompatible with the anchor). | ||
| + | |||
| + | If you want every possible new molecule based on the fragement library you're using, you could set "dn_sampling_method" to "exhaustive" instead of "graph". | ||
| + | |||
| + | ='''Focused ''De Novo'' Design'''= | ||
| + | |||
| + | Focused ''de novo'' design is performed by controlling which moieties can be sampled during the ligand generation process. This is done by generating a fragment library to use instead of the one distributed with DOCK 6.10, and thus controlling the pool of available moieties for DOCK to choose from. Oftentimes the library distributed with DOCK 6.10 is sufficient for most runs, but this methodology can be useful when the generic library lacks a moiety of importance for one's system (such as phosphate), or when the generic library contains moieties that will interact in an undesired way with one's system. | ||
| + | |||
| + | The generation and use of a custom fragment library and the use of the generic fragment library are not mutually exclusive however, and thus this tutorial will also cover how two or more fragment libraries can be combined to make a library containing all of the fragments from both. | ||
| + | |||
| + | =='''Fragment Library Generation'''== | ||
| + | |||
| + | DOCK 6.10 is able to generate a fragment library from an input mol2 file containing one or more ligands. | ||
| + | |||
| + | 1. Move the 3WZE final ligand mol2 file to the focused ''de novo'' directory. We'll be using this file to generate a fragment library, but bear in mind that mol2 files containing multiple ligands can also be used for library generation (and more typically are). | ||
| + | |||
| + | 2. Type "touch frag_gen.in" to make an empty input file for fragment library generation. | ||
| + | |||
| + | 3. Type "dock6 -i frag_gen.in" to go through the question tree presented by DOCK. Use the sample input file below as a guide: | ||
| + | |||
| + | conformer_search_type flex | ||
| + | write_fragment_libraries yes | ||
| + | fragment_library_prefix 3WZE | ||
| + | fragment_library_freq_cutoff 1 | ||
| + | fragment_library_sort_method freq | ||
| + | fragment_library_trans_origin yes | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file 3WZE_final_ligand.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_de_novo.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl | ||
| + | ligand_outfile_prefix trash | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| + | |||
| + | Notes on input parameters: | ||
| + | |||
| + | -'''fragment_library_freq_cutoff''' acts as a sorting mechanism to only allow a fragment into the library if it appears for the specified number of times. When set to 1, this allows any fragment that appears even a single time to be added to the library. | ||
| + | |||
| + | -'''fragment_library_trans_origin''', when set to yes, will translate all fragments in space to a single position, so that when viewed in a program like Chimera or ChimeraX, the user can view the various fragments without having to adjust their camera position each time they switch which fragment that they're looking at. | ||
| + | |||
| + | -'''ligand_outfile_prefix''' is set to "trash" because the output ligand file from fragment library generation will be empty, and can be safely discarded. | ||
| + | |||
| + | 4. After running this fragment library generation, six new files should be produced: | ||
| + | |||
| + | 3WZE_linker.mol2 | ||
| + | 3WZE_rigid.mol2 | ||
| + | 3WZE_scaffold.mol2 | ||
| + | 3WZE_sidechain.mol2 | ||
| + | 3WZE_torenv.dat | ||
| + | trash_scored.mol2 | ||
| + | |||
| + | 5. For this particular fragment library, which was produced only from sorafinib, only the linker, sidechain, and torsions file will contain any information, because sorafinib doesn't contain any groups that DOCK can turn into scaffolds or rigid regions. | ||
| + | |||
| + | 6. Bring the sidechain file to your local machine, and open it with Chimera or ChimeraX. It should contain two sidechains, the first of which should look like the one pictured below: | ||
| + | |||
| + | [[File: fragment1_doug.png|thumb|center|1000px]] | ||
| + | |||
| + | =='''(Optional) Fragment Library Merging'''== | ||
| + | |||
| + | Sometimes, one might wish to combine multiple fragment libraries, and this tutorial will go over combining a fragment library generated by the user, with the one distributed with DOCK 6.10 | ||
| + | |||
| + | 1. Find the fraglib_rigid.mol2, fraglib_scaffold.mol2, fraglib_sidechain.mol2, and fraglib_linker.mol2 files in the parameters/ folder within DOCK6.10/ | ||
| + | |||
| + | 2. Copy them to a the directory where you generated the fragment library for sorafinib. | ||
| + | |||
| + | 3. Type "wc -l fraglib_rigid.mol2" to print the number of lines contained within fraglib_rigid.mol2. Repeat the process for the other fraglib files taken from the parameters/ folder. "wc" stands for word count, and the -l argument makes the command count lines in the input file rather than words. You should find the following values: | ||
| + | |||
| + | 6184 fraglib_linker.mol2 | ||
| + | 706 fraglib_scaffold.mol2 | ||
| + | 10017 fraglib_sidechain.mol2 | ||
| + | 10844 fraglib_torenv.dat | ||
| + | |||
| + | 4. Type "wc -l *3WZE*" to quickly assess how many lines are in each of the files in the library we generated. The output should look like the following: | ||
| + | |||
| + | 229 3WZE_linker.mol2 | ||
| + | 0 3WZE_rigid.mol2 | ||
| + | 0 3WZE_scaffold.mol2 | ||
| + | 60 3WZE_sidechain.mol2 | ||
| + | 6 3WZE_torenv.dat | ||
| + | |||
| + | |||
| + | 5. Type "cat *linker* >> combined_fraglib_linker.mol2" to make a linker file combining the linkers in the generic library and the one generated for sorafinib. Do the same command for the scaffold and sidechain files by substituting the word "linker" for the words "scaffold" and "sidechain" respectively. The "cat" command reads a file and prints its contents, The asterisks are wildcards that ensure that, regardless of the other characters in the filenames, any file with the typed word is taken as an input, the ">>" takes the output of the cat command and adds it to the end of a specified file, which is the empty "combined_fraglib_linker.mol2" in this case. | ||
| + | |||
| + | 5. Locate the "combine_torenv.py" file in the dock6.10/bin/ directory. Copy it over to the directory in which you're generating the combined fragment libraries. | ||
| + | |||
| + | 6. Type "python combine_torenv.py fraglib_torenv.dat 3WZE_torenv.dat". This python script will combine the two torsion files, which cannot simply be combined by appending the contents of two files together as was done in step 4. | ||
| + | |||
| + | 7. To check whether the process was successful, type "wc -l combined_fraglib_*" and repeat the process for the "full_fraglib_torenv.dat" file". Because our library generated from sorafinib only had linkers, sidechains, and torsions, we expect to only see an increase in the number of lines in the full_fraglib_torenv.dat, full_fraglib_linker.mol2, and combined_fraglib_sidechain.mol2 files: | ||
| + | |||
| + | 6413 combined_fraglib_linker.mol2 | ||
| + | 0 combined_fraglib_rigid.mol2 | ||
| + | 6464 combined_fraglib_scaffold.mol2 | ||
| + | 10077 combined_fraglib_sidechain.mol2 | ||
| + | 10850 full_fraglib.dat | ||
| + | |||
| + | The lines in these files are the sums of the files we combined, so this verifies that our fragment libraries have been successfully combined. | ||
| + | |||
| + | =='''Running the Focused ''De Novo'' Design'''== | ||
| + | |||
| + | For this tutorial, we'll use the same 3WZE anchor that we generated for ''de novo'' refinement, and we'll use the fragment library that we generated from sorafinib. | ||
| + | |||
| + | 1. Move the 3WZE anchor mol2 file, and all of the 3WZE fragment library files to a new directory. | ||
| + | |||
| + | 2. Type "touch 3WZE_focused.in" to generate a blank file | ||
| + | |||
| + | 3. Type "dock6 -i 3WZE_focused.in" to go through DOCK's question tree. Use the following input file as a guide for how the questions should be answered: | ||
| + | |||
| + | conformer_search_type denovo | ||
| + | dn_fraglib_scaffold_file 3WZE_scaffold.mol2 | ||
| + | dn_fraglib_linker_file 3WZE_linker.mol2 | ||
| + | dn_fraglib_sidechain_file 3WZE_sidechain.mol2 | ||
| + | dn_user_specified_anchor yes | ||
| + | dn_fraglib_anchor_file Chopped_ligand_for_denovo.mol2 | ||
| + | dn_torenv_table 3WZE_torenv.dat | ||
| + | dn_name_identifier focused_3WZE | ||
| + | dn_sampling_method graph | ||
| + | dn_graph_max_picks 30 | ||
| + | dn_graph_breadth 3 | ||
| + | dn_graph_depth 2 | ||
| + | dn_graph_temperature 100.0 | ||
| + | dn_pruning_conformer_score_cutoff 100.0 | ||
| + | dn_pruning_conformer_score_scaling_factor 2.0 | ||
| + | dn_pruning_clustering_cutoff 100.0 | ||
| + | dn_mol_wt_cutoff_type soft | ||
| + | dn_upper_constraint_mol_wt 550.0 | ||
| + | dn_lower_constraint_mol_wt 0.0 | ||
| + | dn_mol_wt_std_dev 35.0 | ||
| + | dn_constraint_rot_bon 15 | ||
| + | dn_constraint_formal_charge 2.0 | ||
| + | dn_heur_unmatched_num 1 | ||
| + | dn_heur_matched_rmsd 2.0 | ||
| + | dn_unique_anchors 1 | ||
| + | dn_max_grow_layers 1 | ||
| + | dn_max_root_size 25 | ||
| + | dn_max_layer_size 25 | ||
| + | dn_max_current_aps 5 | ||
| + | dn_max_scaffolds_per_layer 1 | ||
| + | dn_write_checkpoints yes | ||
| + | dn_write_prune_dump no | ||
| + | dn_write_orients no | ||
| + | dn_write_growth_trees no | ||
| + | dn_output_prefix 3WZE_focused | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | grid_score_primary yes | ||
| + | grid_score_rep_rad_scale 1 | ||
| + | grid_score_vdw_scale 1 | ||
| + | grid_score_es_scale 1 | ||
| + | grid_score_grid_prefix grid | ||
| + | minimize_ligand no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_de_novo.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl | ||
| + | |||
| + | Note that this process doesn't need to be performed with the anchor we designed for the refinement run. It was only used in this case because it will be easy to visualize the resulting molecules and examine which moieties were added to the single dummy atom. | ||
| + | |||
| + | 4. Import the 3WZE_focused.denovo_build.mol2 file to your local machine, and either use Chimera's viewdock command to open it, or open it in ChimeraX and type "viewdockx". There should be 1 molecule in the file, and it should look like the following: | ||
| + | |||
| + | [[File: focused_doug.png|thumb|center|1000px]] | ||
| − | + | In using sorafinib to generate the library and acnhor, and in only allowing a single growth layer, we've effectively reproduced the original ligand. Obviously, this is not the intended purpose of focused ''de novo'' design, but the lack of numerous output molecules demonstrates that we have successfully performed a ''de novo'' design run with our custom library, otherwise moieties from DOCK 6.10's massive library would have been used to generate numerous ligands, as we observed in the ''de novo' refinement. | |
Latest revision as of 21:42, 7 May 2023
Introduction
This tutorial is a continuation of the virtual screening tutorial. In this tutorial, we'll continue to work with the receptor and ligand in PDB 3WZE, and we'll attempt to generate new ligands for the receptor using three kinds of de novo design: de novo refinement, focused de novo design, and generic de novo design.
De novo can be directly translated as "of new", but a more deft translation might be "from the beginning" or "from scratch". This method of ligand generation involves procedurally generating a ligand using algorithms within programs like DOCK, and is typically used to build entirely new ligands for proteins by building molecules outwards from an initial anchor one moiety at a time.
Generic de novo design best matches the prior description, in which a pre-selected or random anchor is positioned within the active site of the receptor, and then built outwards in a number of layers occupied by various sampled moieties. Focused de novo design is much like generic de novo design, except that the pool of sampled moieties is curtailed to suit the needs of the researcher. Finally, de novo refinement is when one begins with an already discovered ligand, then deletes some of the molecule and replaces it with a dummy atom, effectively using the remainder of the ligand as the anchor for the de novo design algorithms to modify.
Directories
Make new directories for de novo design:
mkdir 009_denovo_generic mkdir 010_denovo_refine mkdir 011_denovo_focused
Generic De Novo Design
In this tutorial, we'll use the generic fragment library distributed with DOCK 6.10, and we won't use a user-specified anchor either. Thus, DOCK will build new ligands for our receptor from scratch.
1. Bring the grid nrg and bmp files for 3WZE's receptor to your current directory. These files are all that you'll need for a generic de novo design run performed within the 3WZE receptor's active site.
2. Type "touch 3WZE_generic.in" to make a blank file.
3. Type "dock6 -i 3WZE_generic.in" to fill out DOCK's question tree and generate an input file. Use the input file below as a guide for how to answer DOCK's questions:
conformer_search_type denovo dn_fraglib_scaffold_file
/gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_scaffold.mol2
dn_fraglib_linker_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_linker.mol2 dn_fraglib_sidechain_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_sidechain.mol2 dn_user_specified_anchor no dn_torenv_table /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_torenv.dat dn_name_identifier 3WZE_generic dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 2.0 dn_pruning_clustering_cutoff 100.0 dn_mol_wt_cutoff_type soft dn_upper_constraint_mol_wt 550.0 dn_lower_constraint_mol_wt 0.0 dn_mol_wt_std_dev 35.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 1 dn_max_grow_layers 9 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_write_checkpoints yes dn_write_prune_dump no dn_write_orients no dn_write_growth_trees no dn_output_prefix 3WZE_generic use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file 3WZE_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter yes bump_grid_prefix grid max_bumps_anchor 2 max_bumps_growth 2 score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix grid minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 250 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_de_novo.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl
4. Terminate the run with control c, then type "nano 3WZE_generic.sh". We're going to run this job on the cluster because it will be too computationally costly for the head node.
5. Copy the following into the file:
#!/bin/bash #SBATCH --job-name=3WZE_generic #SBATCH --ntasks-per-node=24 #SBATCH --nodes=1 #SBATCH --time=48:00:00 #SBATCH -p long-24core dock6 -i 3WZE_generic.in
6. Type "sbatch 3WZE_generic.sh" to start the de novo run on the cluster. The job will probably take 1-2 hours to complete.
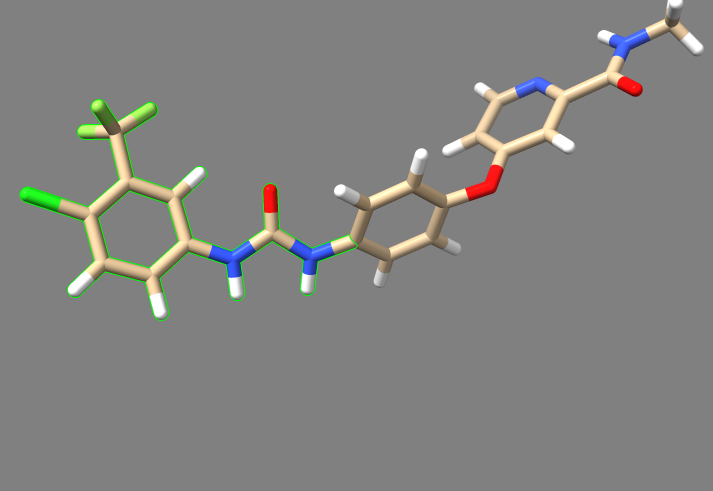
7. Move the "3WZE_generic.denovo_build.mol2" file to your local machine, and either open Chimera and use the viewdock command to open it, or open it in ChimeraX and type "viewdock". There should be approximately 700 ligands produced, and the one with the lowest grid score from our run is shown below:
Rescoring the Outputs
The new ligands generated by this de novo' run have an assocaited grid score to judge their efficacy in binding to the receptor they were designed for, but each ligand can still be energy minimized by rigid docking to arrive at a more accurate estimate of their ability to bind.
1. Type "touch 3WZE_min.in"
2. Type "dock6 -i 3WZE_min.in" and answer the question tree using the following input file as a guide:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file 3WZE_generic.denovo_build.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand yes automated_matching yes receptor_site_file 3WZE_sphere.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter yes bump_grid_prefix grid max_bumps_anchor 2 max_bumps_growth 2 score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix grid minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_de_novo.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix 3WZE_generic_min write_orientations no num_scored_conformers 1 rank_ligands no
3. Bring the "3WZE_generic_min_scored.mol2" file, which contains the minimized molecules to your local machine, and open it with Chimera or ChimeraX as described previously. Shown below is the ligand with the lowest continuous score after energy minmization:
If you compare this ligand to the best scored ligand from the generic de novo design, you'll see that this one actually has a few moieties that are different, suggesting that this molecule is truly best suited for the receptor's binding site, and the prior molecule was only scored highest because all 700 ligands hadn't been energy minimized.
De Novo Refinement
Ligand Preparation
1. Open the final, energy minimized ligand mol2 file which was used for the 3WZE virtual screen tutorial, and also open the final receptor mol2 file that was used in that screen. Either Chimera or ChimeraX can be used to open the files. As long as no translations or rotations have occurred during the virtual screen process, the ligand should still be in its native orientation within the receptor's active site, as depicted by the original 3WZE pdb file.
2. Examine the binding pocket of the receptor, and choose a part of the 3WZE ligand that faces towards the interior of the binding pocket. Parts of the ligand that are innermost to the receptor make for the best parts to delete because they tend to have the most potential interactions with the protein, allowing the various groups tested in de novo design to have a better chance of interacting with a group on the protein. Choosing a part of the ligand to delete which faces the cytosol or the channel leading to the cytosol will be less likely to yield new ligands that can bind tightly to the interior of the receptor. To help recognize good sites for deletion, it's a good idea to show sidechains and hbonds, which can allow you to see which parts of the ligand are interacting with the protein.
In this image, one can see the ligand sorafinib, and also the two hbonds that it forms with the nearby glutamic acid residue 71. It also forms an hbond with the backbone of the receptor using its amide oxygen. Based on this, we'll truncate those two amides and the entire aromatic ring closest to the camera. The camera is positioned to look from the side of the receptor where the binding pocket is deepest, so deleting everything closer than those amides will delete the parts of the ligand which are innermost.
3. Select and delete the receptor. Now that we've identified which part of the ligand to remove, we don't need the receptor anymore.
4. Orient the ligand so that the area you wish to delete is easy to see. Hold control down on your keyboard, then click and drag to cover the area. This should select the area.
5. Now deselect the first atom in the highlighted area. We're going to keep this atom so that it can be changed into a dummy atom. This style of de novo design requires a dummy atom to tell DOCK where to try putting new moieties, and it's easier to keep this nitrogen and change it into a dummy than it is to delete the whole selected area then manually attach a dummy.
6. Delete the selected area using Actions->Atoms/Bonds->Delete. Alternatively, if you're using ChimeraX, simply type "delete sel" into the command line.
You should end up with a molecule that looks like this. Hover your mouse over that nitrogen we spared from deletion, and note its number. In this case the nitrogen is N14.
7. Save this truncated molecule as a mol2 file.
8. Open the mol2 file in a text editor on desktop, or with a command like "nano" from the command line.
9. Find N14, and change the atom type to "Du1". Also change its bond type to "Du". We're only adding a single dummy atom to the anchor in this tutorial because we're trying to modify only one part of an exissting ligand, but bear in mind that one could add any number of dummy atoms to an input ligand and DOCK will try adding moieties to each one.
10. To test whether the mol2 modification worked, open the mol2 with Chimera or ChimeraX. The dummy atom should appear purple or grey, respectively.
(Note that this image was taken with ChimeraX)
11. Now that our ligand is prepared, we can move it to Seawulf where we can perform the actual de novo refinement. For information on how to move a file to Seawulf using the scp command, see the 3WZE virtual screen tutorial.
Running the Refinement
As with the virtual screen, DOCK can be run with an input file, the text of which will be shown below. However, it's a good idea to make your own input file rather than copying what is written here. That way, you can get a sense of what parameters can be adjusted before a de novo refinement run.
1. In the command line in Seawulf, type
touch de_novo_refine.in
Unlike "nano" or "vi", the "touch" command will allow you to make a blank file.
2. To go through the process of answering DOCK's many questions about your run, and to subsequently generate an input file, type
dock6 -i de_novo_refine.in
3. Answer the questions. Our input file is as follows:
conformer_search_type denovo dn_fraglib_scaffold_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_scaffold.mol2 dn_fraglib_linker_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_linker.mol2 dn_fraglib_sidechain_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_sidechain.mol2 dn_user_specified_anchor yes dn_fraglib_anchor_file Chopped_ligand_for_denovo.mol2 dn_torenv_table /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_torenv.dat dn_name_identifier 3WZE_refine dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 2.0 dn_pruning_clustering_cutoff 100.0 dn_mol_wt_cutoff_type soft dn_upper_constraint_mol_wt 1000 dn_lower_constraint_mol_wt 0.0 dn_mol_wt_std_dev 35.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 5 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 1 dn_max_grow_layers 1 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_write_checkpoints yes dn_write_prune_dump no dn_write_orients no dn_write_growth_trees no dn_output_prefix 3WZE_refine use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix grid minimize_ligand yes minimize_anchor no minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_grow_max_iterations 250 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_de_novo.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl
Notes on some of the parameters:
-dn_user_specified_anchor can be set to "no" if one wishes to run a de novo design run in which they do not specify an input anchor with dummy atoms. When set to "no", DOCK 6.10 will use moieties from the fragment libraries as anchors to build out from, allowing for multiple different anchors and orientations to be tried.
-dn_sampling_method can be "graph", "random", or "exhaustive". "graph" will select moieties which are similar to previously selected moieties that improved the grid score, so graph will attempt to bias future moiety selection in a way that promotes improving the ligand. "random" will cause moieties to be selected at random. "exhaustive" will ensure that every possible moiety will be tried at every possible position. Be careful with "exhaustive" though, because it will increase the computation time required in a way proportional to the fragment library size.
-dn_graph_max_picks and dn_num_random_picks (the latter is not shown above) control how many moieties are tried per dummy atom per growth layer.
-dn_max_grow_layers controls how many moieties outwards DOCK will grow your ligand from each dummy atom on the initial anchor. We've only set it to 1 in this refinement tutorial for simplicity, but it can be set higher values. 8 to 9 layers is common for growing ligands from scratch without a set anchor, but the total number of layers that you may want will depend on the goal of the de novo design run.
-minimize_ligand, when set to yes, will attempt to energy minimize the ligand after each moiety selection, in much the same way DOCK 6.10 does so in flexible docking.
-minimize_anchor, when set to yes, will try to energy minimize the anchor before attaching moieties, irrespective of whether the anchor is provided by the user or is chosen randomly. For de novo runs in which the anchor is part of a ligand that binds in a known orientation, it is best to set this parameter to "no". Otherwise, DOCK 6.10 might alter the orientation of your anchor before attempting to grow it with new moieties, and a resulting molecule might not be reflective of how the ligand normally binds.
-Much like the prior parameter, simplex_restraint_min is useful when running a de novo design with a user-supplied anchor of known orientation. When set to yes, this parameter essentially applies a stretchy tether to the initial anchor, allowing it to deviate from its starting position somewhat for the sake of energy minimization, but also applies a penalty based on increasing RMSD.
4. Run the de novo refinenment in Seawulf using the following command:
dock6 -i de_novo_refine.in -o de_novo_refine.out
This should take a few minutes to complete, and when it does, there should be three new files:
3WZE_refine.anchor_1.root_layer_1.mol2 3WZE_refine.denovo_build.mol2 de_novo_refine.out
Checking the Results
1. Bring the two mol2 files to your local machine
2. If you're using Chimera, open Chimera and use Tools->Surface/Binding Analysis->ViewDock to open 3WZE_refine.denovo_build.mol2. If you're using ChimeraX, open that file, then type "viewdockx" into the command line.
Note that this image was taken from ChimeraX. Also, please disregard that I named my ligand "blooble".
3. Look through the results. Because we set "dn_max_grow_layers" to 1, this means that the dummy atom should only be replaced with a single fragment, and there should be no further fragments appended to the single one added.
For our results, we got 10 new molecules. If we had wanted more molecules, we could have increased the value of "dn_graph_max_picks" from 30 to a higher value. This means that dock would sample more fragments (We only got 10 molecules because 20 of the 30 picked fragments must have somehow been incompatible with the anchor).
If you want every possible new molecule based on the fragement library you're using, you could set "dn_sampling_method" to "exhaustive" instead of "graph".
Focused De Novo Design
Focused de novo design is performed by controlling which moieties can be sampled during the ligand generation process. This is done by generating a fragment library to use instead of the one distributed with DOCK 6.10, and thus controlling the pool of available moieties for DOCK to choose from. Oftentimes the library distributed with DOCK 6.10 is sufficient for most runs, but this methodology can be useful when the generic library lacks a moiety of importance for one's system (such as phosphate), or when the generic library contains moieties that will interact in an undesired way with one's system.
The generation and use of a custom fragment library and the use of the generic fragment library are not mutually exclusive however, and thus this tutorial will also cover how two or more fragment libraries can be combined to make a library containing all of the fragments from both.
Fragment Library Generation
DOCK 6.10 is able to generate a fragment library from an input mol2 file containing one or more ligands.
1. Move the 3WZE final ligand mol2 file to the focused de novo directory. We'll be using this file to generate a fragment library, but bear in mind that mol2 files containing multiple ligands can also be used for library generation (and more typically are).
2. Type "touch frag_gen.in" to make an empty input file for fragment library generation.
3. Type "dock6 -i frag_gen.in" to go through the question tree presented by DOCK. Use the sample input file below as a guide:
conformer_search_type flex write_fragment_libraries yes fragment_library_prefix 3WZE fragment_library_freq_cutoff 1 fragment_library_sort_method freq fragment_library_trans_origin yes use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file 3WZE_final_ligand.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_de_novo.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix trash write_orientations no num_scored_conformers 1 rank_ligands no
Notes on input parameters:
-fragment_library_freq_cutoff acts as a sorting mechanism to only allow a fragment into the library if it appears for the specified number of times. When set to 1, this allows any fragment that appears even a single time to be added to the library.
-fragment_library_trans_origin, when set to yes, will translate all fragments in space to a single position, so that when viewed in a program like Chimera or ChimeraX, the user can view the various fragments without having to adjust their camera position each time they switch which fragment that they're looking at.
-ligand_outfile_prefix is set to "trash" because the output ligand file from fragment library generation will be empty, and can be safely discarded.
4. After running this fragment library generation, six new files should be produced:
3WZE_linker.mol2 3WZE_rigid.mol2 3WZE_scaffold.mol2 3WZE_sidechain.mol2 3WZE_torenv.dat trash_scored.mol2
5. For this particular fragment library, which was produced only from sorafinib, only the linker, sidechain, and torsions file will contain any information, because sorafinib doesn't contain any groups that DOCK can turn into scaffolds or rigid regions.
6. Bring the sidechain file to your local machine, and open it with Chimera or ChimeraX. It should contain two sidechains, the first of which should look like the one pictured below:
(Optional) Fragment Library Merging
Sometimes, one might wish to combine multiple fragment libraries, and this tutorial will go over combining a fragment library generated by the user, with the one distributed with DOCK 6.10
1. Find the fraglib_rigid.mol2, fraglib_scaffold.mol2, fraglib_sidechain.mol2, and fraglib_linker.mol2 files in the parameters/ folder within DOCK6.10/
2. Copy them to a the directory where you generated the fragment library for sorafinib.
3. Type "wc -l fraglib_rigid.mol2" to print the number of lines contained within fraglib_rigid.mol2. Repeat the process for the other fraglib files taken from the parameters/ folder. "wc" stands for word count, and the -l argument makes the command count lines in the input file rather than words. You should find the following values:
6184 fraglib_linker.mol2 706 fraglib_scaffold.mol2 10017 fraglib_sidechain.mol2 10844 fraglib_torenv.dat
4. Type "wc -l *3WZE*" to quickly assess how many lines are in each of the files in the library we generated. The output should look like the following:
229 3WZE_linker.mol2 0 3WZE_rigid.mol2 0 3WZE_scaffold.mol2 60 3WZE_sidechain.mol2 6 3WZE_torenv.dat
5. Type "cat *linker* >> combined_fraglib_linker.mol2" to make a linker file combining the linkers in the generic library and the one generated for sorafinib. Do the same command for the scaffold and sidechain files by substituting the word "linker" for the words "scaffold" and "sidechain" respectively. The "cat" command reads a file and prints its contents, The asterisks are wildcards that ensure that, regardless of the other characters in the filenames, any file with the typed word is taken as an input, the ">>" takes the output of the cat command and adds it to the end of a specified file, which is the empty "combined_fraglib_linker.mol2" in this case.
5. Locate the "combine_torenv.py" file in the dock6.10/bin/ directory. Copy it over to the directory in which you're generating the combined fragment libraries.
6. Type "python combine_torenv.py fraglib_torenv.dat 3WZE_torenv.dat". This python script will combine the two torsion files, which cannot simply be combined by appending the contents of two files together as was done in step 4.
7. To check whether the process was successful, type "wc -l combined_fraglib_*" and repeat the process for the "full_fraglib_torenv.dat" file". Because our library generated from sorafinib only had linkers, sidechains, and torsions, we expect to only see an increase in the number of lines in the full_fraglib_torenv.dat, full_fraglib_linker.mol2, and combined_fraglib_sidechain.mol2 files:
6413 combined_fraglib_linker.mol2 0 combined_fraglib_rigid.mol2 6464 combined_fraglib_scaffold.mol2 10077 combined_fraglib_sidechain.mol2 10850 full_fraglib.dat
The lines in these files are the sums of the files we combined, so this verifies that our fragment libraries have been successfully combined.
Running the Focused De Novo Design
For this tutorial, we'll use the same 3WZE anchor that we generated for de novo refinement, and we'll use the fragment library that we generated from sorafinib.
1. Move the 3WZE anchor mol2 file, and all of the 3WZE fragment library files to a new directory.
2. Type "touch 3WZE_focused.in" to generate a blank file
3. Type "dock6 -i 3WZE_focused.in" to go through DOCK's question tree. Use the following input file as a guide for how the questions should be answered:
conformer_search_type denovo dn_fraglib_scaffold_file 3WZE_scaffold.mol2 dn_fraglib_linker_file 3WZE_linker.mol2 dn_fraglib_sidechain_file 3WZE_sidechain.mol2 dn_user_specified_anchor yes dn_fraglib_anchor_file Chopped_ligand_for_denovo.mol2 dn_torenv_table 3WZE_torenv.dat dn_name_identifier focused_3WZE dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 2.0 dn_pruning_clustering_cutoff 100.0 dn_mol_wt_cutoff_type soft dn_upper_constraint_mol_wt 550.0 dn_lower_constraint_mol_wt 0.0 dn_mol_wt_std_dev 35.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 1 dn_max_grow_layers 1 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_write_checkpoints yes dn_write_prune_dump no dn_write_orients no dn_write_growth_trees no dn_output_prefix 3WZE_focused use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix grid minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_de_novo.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl
Note that this process doesn't need to be performed with the anchor we designed for the refinement run. It was only used in this case because it will be easy to visualize the resulting molecules and examine which moieties were added to the single dummy atom.
4. Import the 3WZE_focused.denovo_build.mol2 file to your local machine, and either use Chimera's viewdock command to open it, or open it in ChimeraX and type "viewdockx". There should be 1 molecule in the file, and it should look like the following:
In using sorafinib to generate the library and acnhor, and in only allowing a single growth layer, we've effectively reproduced the original ligand. Obviously, this is not the intended purpose of focused de novo design, but the lack of numerous output molecules demonstrates that we have successfully performed a de novo design run with our custom library, otherwise moieties from DOCK 6.10's massive library would have been used to generate numerous ligands, as we observed in the de novo' refinement.