Difference between revisions of "2012 AMBER Tutorial with Biotin and Streptavidin"
Stonybrook (talk | contribs) (→MMGBSA Energy Calculation) |
(→Plotting your results) |
||
| (27 intermediate revisions by one other user not shown) | |||
| Line 393: | Line 393: | ||
===Ptraj=== | ===Ptraj=== | ||
| − | |||
To run the Ptraj analysis, you will probably first create a separate folder for it under the root directory if you don't have one. | To run the Ptraj analysis, you will probably first create a separate folder for it under the root directory if you don't have one. | ||
mkdir 004.PTRAJ | mkdir 004.PTRAJ | ||
| Line 423: | Line 422: | ||
ptraj ../002.ANTE.TLEAP/1DF8.com.gas.leap.parm ptraj.3.in > ptraj.3.log | ptraj ../002.ANTE.TLEAP/1DF8.com.gas.leap.parm ptraj.3.in > ptraj.3.log | ||
| − | |||
Finally, you want to perform MMGBSA analysis to obtain energetic information about the system. To do this, you need a trajectory file of the gas phase complex (which we already created above and saved as 1DF8.com.trj.stripfit), and want to create two more trajectory files containing just the receptor and just the ligand. To do so, you have to run two more ptraj using the following ptraj input files: | Finally, you want to perform MMGBSA analysis to obtain energetic information about the system. To do this, you need a trajectory file of the gas phase complex (which we already created above and saved as 1DF8.com.trj.stripfit), and want to create two more trajectory files containing just the receptor and just the ligand. To do so, you have to run two more ptraj using the following ptraj input files: | ||
| Line 456: | Line 454: | ||
In the snapshot above, the water molecules are shown as points. | In the snapshot above, the water molecules are shown as points. | ||
| + | |||
| + | ====RMSD Plots==== | ||
| + | Once you have finished running Ptraj, you should have two RMSD files: 1df8.lig.rmsd.txt 1df8.rmsd.CA.txt | ||
| + | |||
| + | The data in these files can be plotted to visualize the structural drift (ie change in RMSD) over the course of your simulation. Large fluctuations in RMSD may be an indication that your simulation is unstable. | ||
| + | |||
| + | Here is an example plot for ligand RMSD: | ||
| + | |||
| + | [[Image:2012_AMBER_RMSD_plot.jpg|500px|center]] | ||
| + | |||
| + | ====Measuring h-bond distances==== | ||
| + | |||
| + | Ptraj can also be used to measure the hydrogen bonding distance between two atoms. To use this functionality, we first need to decide which atom pairs are forming interesting h-bonds. We can do this with VMD, which can draw H-bonds automatically for us. First, select the ligand (BTN) and residues around the ligand. Second, change the '''Drawing Method''' to '''HBonds'''. | ||
| + | |||
| + | After you have picked out some atom pairs, use the '''distance''' command in Ptraj to measure the distance between the two atoms. | ||
| + | |||
| + | Here is a sample script: | ||
| + | |||
| + | #! /bin/tcsh | ||
| + | #PBS -l nodes=1:ppn=1 | ||
| + | #PBS -l walltime=01:00:00 | ||
| + | #PBS -o zzz.qsub.out | ||
| + | #PBS -e zzz.qsub.err | ||
| + | #PBS -V | ||
| + | set workdir = "~/AMBER_tutorial/007.PTRAJ.HBONDS" | ||
| + | cd ${workdir} | ||
| + | ptraj ../002.TLEAP/1df8.com.gas.leap.prm7 << EOF | ||
| + | trajin ../004.PTRAJ/1df8.com.trj.stripfit | ||
| + | distance 34N_119O2 :34@N :119@O2 out 34N_119O2.out | ||
| + | distance 73OG_119O3 :73@OG :119@O3 out 73OG_119O3.out | ||
| + | distance 12OG_119O14 :12@OG :119@O14 out 12OG_119O14.out | ||
| + | distance 28OH_119O14 :28@OH :119@O14 out 28OH_119O14.out | ||
| + | EOF | ||
| + | |||
| + | The results can be easily plotted by writing a script for XMGRACE. Here is an example: | ||
| + | |||
| + | #!/bin/tcsh | ||
| + | foreach distance (28OH_119O14 12OG_119O14) | ||
| + | cat << EOF > $distance.grace.in | ||
| + | READ NXY "$distance.out" | ||
| + | s0 legend "$distance" | ||
| + | title "Example Hydrogen Bond Distance Plot" | ||
| + | xaxis label "Time (picoseconds)" | ||
| + | yaxis label "Hydrogen Bond Distance (Angstroms)" | ||
| + | PRINT TO "$distance.eps" | ||
| + | DEVICE "EPS" OP "level2" | ||
| + | PRINT | ||
| + | EOF | ||
| + | xmgrace -batch $distance.grace.in -nosafe -hardcopy | ||
| + | end | ||
| + | |||
| + | This will generate two plots for the above hydrogen bond distances in EPS format, here is one of them: | ||
| + | |||
| + | [[Image:2012_AMBER_hbond_plot.jpg|500px|center]] | ||
===MMGBSA Energy Calculation=== | ===MMGBSA Energy Calculation=== | ||
| − | '''MM/GBSA''' is the acronym for '''Molecular Mechanics/ | + | '''MM/GBSA''' is the acronym for '''Molecular Mechanics/Generalized Born Surface Area'''. This part of '''AMBER''' combines molecular mechanics ('''MM''') with both the electrostatic ('''PB''') and nonpolar ('''SA''') contribution to solvation . Topology files are needed for the receptor ('''R'''), ligand ('''L'''), and receptor-ligand complex ('''RL'''). The trajectory files ('''trj''') generate the coordinates. Therefore, molecular dynamics is used to generate a set of snapshots taken at fixed intervals from the trajectories. These snapshots are processed to remove solvent and generate the free energy of binding. |
In the '''AMBER_Tutorial''' directory, create a new directory: | In the '''AMBER_Tutorial''' directory, create a new directory: | ||
| Line 465: | Line 517: | ||
mkdir 005.MMGBSA | mkdir 005.MMGBSA | ||
| − | In this new directory, create the file '''gb. | + | In this new directory, create the file '''gb.rescore.in''' containing: |
Single point GB energy calc | Single point GB energy calc | ||
| Line 554: | Line 606: | ||
'''grep ESURF gb.rescore.out.com > surf.com.txt'''. | '''grep ESURF gb.rescore.out.com > surf.com.txt'''. | ||
| + | You can take these text files, import them into Excel, and do the rest of your modifications there. | ||
| + | |||
| + | ====Equations for analysis==== | ||
| + | |||
| + | Remember that to obtain the ''Gvdw'' term, you need to take the ''SASA'' (which is ESURF) and input into equation 1: | ||
| + | |||
| + | ''ΔGnonpolar'' = ''SASA''*0.00542 + 0.92 [1] | ||
| + | |||
| + | Also, the mmgbsa of a given system can be determined by equation 2: | ||
| + | |||
| + | ''ΔGmmgbsa'' = ''ΔGvdw'' + ''ΔGcoul'' + ''ΔGpolar'' + ''ΔGnonpolar'' [2] | ||
| + | |||
| + | From the output file: | ||
| − | ===Plotting your results=== | + | VDWAALS = ''ΔGvdw'' |
| + | |||
| + | EELS = ''ΔGcoul'' | ||
| + | |||
| + | EGB = ''ΔGpolar'' | ||
| + | |||
| + | You can then easily calculate the ΔΔGbind by using equation 3: | ||
| + | |||
| + | ''ΔΔGbind = ΔGmmgbsa,complex – (ΔGmmgbsa,lig + ΔGmmgbsa,rec)'' [3] | ||
| + | |||
| + | You will want to careful when doing your analysis that the results from frame 1 for the receptor and ligand are subtracted from the results from frame 1 for your complex. By doing this in excel, you should have 2000 frames for each, and the values should cleanly line up. Finally, you will want to plot your ''ΔΔGbind'' and examine if you see corresponding changes in the ligand position and the ''ΔΔGbind''. Also, you should determine the mean and standard deviation for your ''ΔΔGbind''. | ||
| + | |||
| + | ====Plotting your results==== | ||
When your rescoring calculation finishes, you have the following three output files: "gb.rescore.out.com", "gb.rescore.out.lig", and "gb.rescore.out.rec". | When your rescoring calculation finishes, you have the following three output files: "gb.rescore.out.com", "gb.rescore.out.lig", and "gb.rescore.out.rec". | ||
| Line 570: | Line 647: | ||
grep VDWAALS gb.rescore.out.$i | awk '{print $3}' > $i.vdw | grep VDWAALS gb.rescore.out.$i | awk '{print $3}' > $i.vdw | ||
| − | grep | + | grep EGB gb.rescore.out.$i | awk '{print $9}' > $i.polar |
| − | grep | + | grep EELS gb.rescore.out.$i | awk '{print $6}' > $i.coul |
grep ESURF gb.rescore.out.$i | awk '{print $3 * 0.00542 + 0.92}' > $i.surf | grep ESURF gb.rescore.out.$i | awk '{print $3 * 0.00542 + 0.92}' > $i.surf | ||
| Line 599: | Line 676: | ||
bash get.mmgbsa.sh | bash get.mmgbsa.sh | ||
| − | This will create a text file called MMGBSA_vs_time with x and y values separated by a space and comma. These values can be imported to Excel or Origin or to XMGRACE if you are using Linux: | + | This will create a text file called MMGBSA_vs_time.dat with x and y values separated by a space and comma. These values can be imported to Excel or Origin or to XMGRACE if you are using Linux: |
xmgrace MMGBSA_vs_time.dat | xmgrace MMGBSA_vs_time.dat | ||
| − | + | [[Image:2012 AMBER MMGBSA plot.jpg|500px|center]] | |
| − | |||
| − | [[Image:2012 AMBER MMGBSA plot.jpg| | ||
==V. Frequently Encountered Problems== | ==V. Frequently Encountered Problems== | ||
| Line 659: | Line 734: | ||
In structural preparation step, you should treat both proteins as receptors. Remove the water molecules and other small molecules needed. (Do not add hydrogens and charges in this case). Save the file into '''pdb''' fomat as Woo Suk wrote. You may need to modify the some pdb files. E.g. change residue name '''CYS''' to '''CYX''', change '''SH''' to '''SG''' otherwise it will not be recognized by tleap. | In structural preparation step, you should treat both proteins as receptors. Remove the water molecules and other small molecules needed. (Do not add hydrogens and charges in this case). Save the file into '''pdb''' fomat as Woo Suk wrote. You may need to modify the some pdb files. E.g. change residue name '''CYS''' to '''CYX''', change '''SH''' to '''SG''' otherwise it will not be recognized by tleap. | ||
| + | |||
| + | '''Plot per-residue RMSF (Root mean square fluctuation) value to 3D structure''' | ||
| + | |||
| + | Generate RMSF values using ptraj: | ||
| + | creat ptraj.in in vim. Write scripts like this. | ||
| + | trajin 1G9M.com.trj.stripfit 1 2000 1 | ||
| + | reference ../002.ANTE.TLEAP/1G9M.com.gas.leap.rst7 | ||
| + | average avg.pdb pdb | ||
| + | atomicfluct out RMSF.dat :1-486@CA byres | ||
| + | run ptraj: | ||
| + | ptraj ../002.ANTE.TLEAP/1G9M.com.gas.leap.prm7 ptraj.in > ptraj.log | ||
| + | you will get two output files: avg.pdb and RMSF.dat. | ||
| + | You can generate B factor data instead of RMSF by adding "bfactor" to the end of last line. To calculate any other properties, you can refer to the AMBER | ||
| + | Tools User's Manual. | ||
| + | |||
| + | |||
| + | Map RMSF to 3D structure: | ||
| + | copy the second column of RMSF.dat file to the last column (beta column) of avg.pdb. You can make a "perl" script, or use excel to do that. In my case, I type "vimdiff avg.pdb RMSF.dat" to open two files together. Then use visual block (ctrl + v) to copy (y) and paste (p). | ||
| + | You can copy any column of per-residue properties to the beta column of pdb file you care about. E.g. RMSD, energy, footprint interactions, sequence conservation. | ||
| + | |||
| + | Then you can use vmd to draw the plot: VMD Main --> Graphics --> Representation --> Coloring Method --> Beta. | ||
| + | You can change the color scale: VMD Main --> Graphics --> color --> Color Scale --> Method --> whatever color range you like. | ||
===Yuchen=== | ===Yuchen=== | ||
| + | One important thing is that you should always make sure that you have the right ligand, receptor and complex structure before you run Ptraj. Because Ptraj is a relatively expensive calculation, for example in this tutorial, the calculation takes around 80 hours when using 8 processors for parallel computing, you don't want to find out that your results are not right simply because you have a typo in your TLEAP input file. What you can do is that after you generate the parameter and restart file using TLEAP, you can use VMD to visually check if you have the right structure. | ||
===Yunting=== | ===Yunting=== | ||
| Line 690: | Line 788: | ||
Note that .crd files are the same as .rst7 -- you can simply change the extension if you wish. | Note that .crd files are the same as .rst7 -- you can simply change the extension if you wish. | ||
| − | ==== | + | ====Simulation with explicit solvent==== |
| − | Note that if you have | + | Note that if you have (explicit) waters in your AMBER simulation, you will need to use a periodic box |
| + | |||
| + | ====XMGRACE on Mac OS==== | ||
| + | You'll need to install the OpenMotif libraries first: http://www.ist.co.uk/downloads/motif_download.html | ||
| + | |||
| + | Then do: | ||
| + | |||
| + | cd Desktop | ||
| + | wget ftp://ftp.fu-berlin.de/unix/graphics/grace/src/grace5/grace-latest.tar.gz | ||
| + | tar xzvf grace-latest.tar.gz | ||
| + | cd grace-5.1.22 | ||
| + | ./configure --x-includes=/usr/X11/include --x-libraries=/usr/X11/lib \ | ||
| + | --with-motif-library=-lXm --with-extra-incpath=/usr/OpenMotif/include:/Users/sm4082/fftw-2.1.5/include \ | ||
| + | --with-extra-ldpath=/usr/OpenMotif/lib:/Users/sm4082/fftw-2.1.5/lib | ||
| + | make | ||
| + | sudo make install | ||
| + | echo "export PATH=$PATH:/usr/local/grace/bin" >> ~/.bashrc | ||
| + | source ~/.bashrc | ||
| + | xmgrace | ||
| + | |||
| + | Alternatively, you can also install XMGRACE using [http://www.finkproject.org/ Fink] or [http://www.macports.org/ MacPorts] | ||
| + | |||
| + | ====Running out of disk space==== | ||
| + | If you fill up your quota of disk space on Herbie, you can temporarily store files in the scratch space located at '''/space1''' | ||
| + | |||
| + | Please note that /space1 is for ''temporary'' storage only and this space is periodically erased. Nevertheless, this scratch space is very useful for moving files from Seawulf to Herbie and then to your personal computer for permanent storage (or for temporarily storing files that you want to visualize on a math lab computer, etc.) | ||
Latest revision as of 10:31, 26 March 2014
For additional Rizzo Lab tutorials see AMBER Tutorials.
In this tutorial, we will learn how to run a molecular dynamics simulation of a protein-ligand complex. We will then post-process that simulation by calculating structural fluctuations (with RMSD) and free energies of binding (MM-GBSA).
Contents
I. Introduction
AMBER
Amber - Assisted Model Building with Energy Refinement - is a suite of about multiple programs for perform macromolecular simulations. Amber11, the current version of Amber, includes newly released functionality such as PMEMD, particle mesh Ewald MD and soft-core Thermodynamics Integration MD. For the tutorial, we are using an older version which is AMBER10.
The Amber 10 Manual is the primary resource to get started with Amber10. (Tip: Using Adobe Acrobat to view the file, you can simply search the document for keywords such as the name of a simulation parameter, which saves much time.) In addition, Amber Tools User's Manual serves as another reference while using Amber tools. You can also read the manual for Amber11 on Amber11 and AmberTools Users' Manuals
Here are some programs in Amber
- LEaP: an preparing program for constructing new or modified systems in Amber. It consists of the functions of prep, link, edit, and parm for earlier version of Amber.
- ANTECHAMBER: in additional to LEap, this main Antechamber suite program is for preparing input files other than standard nucleic acids and proteins.
- SANDER: according to the Amber 10 manual, it is 'a basic energy minimizer and molecular dynamics program' that can be used to minimize, equilibrate and sample molecular conformations. And this is the program we mainly use in this tutorial to generate trajectory files of the molecular system.
- PMEMD: version of SANDER that has improved parallel scaling property and optimized speed.
- PTRAJ: an analysis program for processing trajectory files. One can use ptraj to rotate, translate the structures, evaluate geometrical features and so on.
There is a mailing list you could sign-up for, as an additional resource.
Biotin and Streptavidin
For information of the Biotin and Streptavidin system, see 2012 DOCK tutorial with Streptavidin.
Organizing Directories
While performing MD simulations, it is convenient to adopt a standard directory structure / naming scheme, so that files are easy to find / identify. For this tutorial, we will use something similar to the following:
~username/AMS536/AMBER-Tutorial/001.CHIMERA.MOL.PREP/
002.ANTE.TLEAP/
003.SANDER/
004.PTRAJ/
005.MMGBSA/
II. Structural Preparation
Preparation in Chimera
In this AMBER tutorial, we will use the same system with previous DOCK part. Chimera can directly get the structure by its PDB ID 1DF8. To begin with, we need three files under directory 001.CHIMERA.MOL.PREP.
1DF8.rec.lig.pdb:
1. To lower computational cost and make the system clear, we remove chain B of the dimer. Select - chain - B, Action - Atoms/Bonds - delete
2. Remove the water molecules. Select - residue - HOH, Action - Atoms/Bonds - delete
Then we separate the receiver and its ligand.
1DF8.rec.noH.pdb:
Select the ligand and delete it. Select - Residue - BTN, Action - Atoms/Bonds - delete
1DF8.lig.chimera.mol2:
1. Select the protein and delete it. Select - Residue - BTN, Select - Invert, Action - Atoms/Bonds - delete
2. Use Dock Prep to add hydrogens and charges(AM1-BCC) to the ligand. Tools - Structure Editing - Dock Prep
antechamber
An antechamber input file requires all the atom names to be unique and it only uses the first 3 characters as the name. So if we use 1DF8.lig.chimera.mol2 as the input file, it will cause errors("H102" and "H103" will have the same name "H10").
14 O3 26.9770 10.6020 12.2050 O.2 1 BTN201 -0.6531
15 N2 28.6480 12.1210 11.8210 N.pl3 1 BTN201 -0.4789
16 C4 28.9670 13.2060 10.9010 C.3 1 BTN201 0.0816
17 H102 32.0358 17.4077 15.9597 H 1 BTN201 0.0272
18 H103 30.6885 18.0473 15.0102 H 1 BTN201 0.0219
19 H92 30.5603 15.6243 15.3130 H 1 BTN201 -0.0094
20 H93 32.1288 15.4384 14.4982 H 1 BTN201 0.0384
21 H82 29.6624 16.7125 13.2433 H 1 BTN201 0.0275
We need to manually rename the atoms. One way is to use the first column numbers to be the atom names. If you are using Vim, the visual block mode can help by selecting a rectangular section of text. We rename the atoms and save the file as 1DF8.lig.mol2 under directory 001.CHIMERA.MOL.PREP.
14 O14 26.9770 10.6020 12.2050 O.2 1 BTN201 -0.6531
15 N15 28.6480 12.1210 11.8210 N.pl3 1 BTN201 -0.4789
16 C16 28.9670 13.2060 10.9010 C.3 1 BTN201 0.0816
17 H17 32.0358 17.4077 15.9597 H 1 BTN201 0.0272
18 H18 30.6885 18.0473 15.0102 H 1 BTN201 0.0219
19 H19 30.5603 15.6243 15.3130 H 1 BTN201 -0.0094
20 H20 32.1288 15.4384 14.4982 H 1 BTN201 0.0384
21 H21 29.6624 16.7125 13.2433 H 1 BTN201 0.0275
LEaP
To begin with, go to 002.ANTE.TLEAP directory.
To make sure we have access to the three programs that we want to run (antechamber, parmchk and tleap) and we are using the correct version of amber, we can use the which command, type:
which antechamber which parmchk which tleap
You should have results similar to this:
/nfs/user03/tbalius/amber10_ompi/bin/antechamber /nfs/user03/tbalius/amber10_ompi/bin/parmchk /nfs/user03/tbalius/amber10_ompi/bin/tleap
Please note that you need to be in the tcsh shell for running the which command shown above. Also, if you are using a different version of amber, you might want to change to the correct version by editing the .cshrc file and source it.
Copy parameters of ions to your working directory from the following resource:
scp -r ~lingling/AMS536/AMBER_Tutorial/002.ANTE.TLEAP/rizzo_amber7.ionparms .
Antechamber is a set of tools to generate files for organic molecules, which can then be read into LEaP. The antechamber program itself is the main program of Antechamber package. It can perform many file conversions, and can also assign atomic charges and atom types. In this tutorial, we use antechamber to convert our input mol2 file into files ready for LEaP. In the command line, type:
antechamber -i ../001.CHIMERA.MOL.PREP/1DF8.lig.mol2 -fi mol2 -o 1DF8.lig.ante.pdb -fo pdb
-i input file name; -fi input file format; -o output file name; -fo output file format. You will have an output file:
1DF8.lig.ante.pdb
Similarly, we can use antechamber to change the fomat of 1DF8.lig.mol2 file to prep file:
antechamber -i ../001.CHIMERA.MOL.PREP/1DF8.lig.mol2 -fi mol2 -o 1DF8.lig.ante.prep -fo prepi
You will get a set of output files:
ANTECHAMBER_BOND_TYPE.AC ATOMTYPE.INF 1DF8.lig.ante.prep ANTECHAMBER_BOND_TYPE.AC0 NEWPDB.PDB ANTECHAMBER_AC.AC ANTECHAMBER_PREP.AC PREP.INF ANTECHAMBER_AC.AC0 ANTECHAMBER_PREP.AC0
These files give detailed information of the ligand such as atom coordinates, bond angle, bond length, dihedral angles, etc. For example:
vim ATOMTYPE.INF
You can see this file contains information related to the ring system.
Parmchk is another program included in the Antechamber package. After running antechamber, we run parmchk to check the parameters. If there are missing parameters after antechamber is finished, the frcmod template generated by parmchk will help us in generating the needed parameters:
parmchk -i 1DF8.lig.ante.prep -f prepi -o 1DF8.lig.ante.frcmod
Then open the output file 1DF8.lig.ante.frcmod, you will see something like this:
remark goes here MASS BOND ANGLE DIHE IMPROPER c3-o -c -o 1.1 180.0 2.0 General improper torsional angle (1 general atom type) n -n -c -o 10.5 180.0 2.0 General improper torsional angle (2 general atom types) NONBON
This means all the needed force field parameters are avalaible except there are two missing dihedral parameters, which were assigned a default value.
Next, we need create 3 new input files – “tleap.lig.in”, “tleap.rec.in” and “tleap.com.in”, for the ligand, the receptor, and the complex, respectively. These input files will be used by LEaP to create parameter/topology files and initial coordinate files.
vim tleap.lig.in
In this new ligand input file, type:
set default PBradii mbondi2 #set default PBradii source leaprc.ff99SB #load a force field loadoff rizzo_amber7.ionparms/ions.lib loadamberparams rizzo_amber7.ionparms/ions.frcmod #load an AMBER format parameter file source leaprc.gaff loadamberparams rizzo_amber7.ionparms/gaff.dat.rizzo loadamberparams 1DF8.lig.ante.frcmod loadamberprep 1DF8.lig.ante.prep #load an AMBER PREP file lig = loadpdb 1DF8.lig.ante.pdb #load the ligand pdb file saveamberparm lig 1DF8.lig.gas.leap.prm7 1DF8.lig.gas.leap.rst7 #save the ligand gas phase AMBER topology and coordinate files solvateBox lig TIP3PBOX 10.0 #solvate the receptor using TIP3P, solvent box radii 10 angstroms saveamberparm lig 1DF8.lig.wat.leap.prm7 1DF8.lig.wat.leap.rst7 #save the ligand water phase AMBER topology and coordinate files charge lig #charge the ligand quit
Similarly, create the tleap.rec.in and tleap.com.in files:
set default PBradii mbondi2 source leaprc.ff99SB loadoff rizzo_amber7.ionparms/ions.lib loadamberparams rizzo_amber7.ionparms/ions.frcmod REC = loadpdb ../001.CHIMERA.MOL.PREP/1DF8.rec.noh.pdb saveamberparm REC 1DF8.rec.gas.leap.prm7 1DF8.rec.gas.leap.rst7 solvateBox REC TIP3PBOX 10.0 saveamberparm REC 1DF8.rec.wat.leap.prm7 1DF8.rec.wat.leap.rst7 charge REC quit
set default PBradii mbondi2
source leaprc.ff99SB
loadoff rizzo_amber7.ionparms/ions.lib
loadamberparams rizzo_amber7.ionparms/ions.frcmod
source leaprc.gaff
loadamberparams rizzo_amber7.ionparms/gaff.dat.rizzo
REC = loadpdb ../001.CHIMERA.MOL.PREP/1DF8.rec.noh.pdb
loadamberparams 1DF8.lig.ante.frcmod
loadamberprep 1DF8.lig.ante.prep
LIG = loadpdb 1DF8.lig.ante.pdb
COM = combine {REC LIG}
saveamberparm COM 1DF8.com.gas.leap.prm7 1DF8.com.gas.leap.rst7
solvateBox COM TIP3PBOX 10.0
saveamberparm COM 1DF8.com.wat.leap.prm7 1DF8.com.wat.leap.rst7
charge COM
quit
Use tleap command:
tleap -s -f tleap.lig.in > tleap.lig.out tleap -s -f tleap.rec.in > tleap.rec.out tleap -s -f tleap.com.in > tleap.com.out
you will see the following output files:
1DF8.lig.gas.leap.prm7 1DF8.lig.gas.leap.rst7 1DF8.lig.wat.leap.prm7 1DF8.lig.wat.leap.rst7 1DF8.rec.gas.leap.prm7 1DF8.rec.gas.leap.rst7 1DF8.rec.wat.leap.prm7 1DF8.rec.wat.leap.rst7 1DF8.com.gas.leap.prm7 1DF8.com.gas.leap.rst7 1DF8.com.wat.leap.prm7 1DF8.com.wat.leap.rst7 tleap.lig.out tleap.rec.out tleap.com.out leap.log
An alternative way to use antechamber, parmchk and tleap is to make a csh script, which contains the commands you want to do, and run the script.
You can visualize the output files in VMD, and use the output files to run SANDER.
Reference:
2011 AMBER Tutorial with Biotin and Streptavidin
Visualization in VMD
First, copy “002.ANTE.TLEAP”directory back to Herbie. On Herbie,type the following commands:
cd ~/AMS536/AMBER_Tutorial/ scp –r sw:~/AMS536/AMBER_Tutorial/002.ANTE.TLEAP/ .
Open VMD software:
vmd
Then, we can load the gas phase ligand file generated by tleap: VMD Main->File->New Molecule (Please note that in order to visualize in VMD, we need first load the .prm7 file and then load the .rst7 file)
Load file for: New Molecule File name: 1DF8.lig.gas.leap.prm7 Determine file type: AMBER7 Parm
Load file for: 1DF8.lig.gas.leap.prm7 File name: 1DF8.lig.gas.leap.rst7 Determine file type: AMBER7 Restart
You can see the ligand in the gas phase looks like this:
You can compare this with ligand in the water phase by opening 1DF8.lig.water.leap.prm7, 1DF8.lig.water.leap.rst7 at the same time.
In order to see the ligands clearly, we can visualize without hydrogen atoms. VMD Main->Graphics->Representations->
In the Selected Atoms command line, type: all and noh->Click Apply
You can actually see the ligand in water is very different from that in gas phase.
III. Simulation using sander
Minimization
Before we actually run the molecular dynamics simulation, an energy minimization step must be performed. The purpose of this step is to remove structural artifacts resulting from the model-building process. The most common approach to energy minimization is the “positional restraint” method, which essential works by tying each atom in the model to a reference position with a weak spring. In each cycle of minimization, the atoms in the model will move some distance away from the reference positions to which they are restrained, and the new positions they occupy will be used as the reference for the next cycle. After multiple cycles, the conformation of the model will approach a local energy minimum where the positions of the atoms will not further change.
Listed below is a typical input file “01mi.in” for minimization:
01mi.in: equilibration &cntrl imin = 1, maxcyc = 1000, ntmin = 2, ntx = 1, ntc = 1, ntf = 1, ntb = 1, ntp = 0, ntwx = 1000, ntwe = 0, ntpr = 1000, scee = 1.2, cut = 8.0, ntr = 1, restraintmask = ':1-119 & !@H=', restraint_wt=5.0, /
In the input file, “imin = 1” specifies that minimization instead of molecular dynamics is being performed here. The parameter “maxcyc” specifies the total number of minimization cycles we will run, while “ntmin” specifies how many of those cycles will be run using the deepest descent method. Here a total of 1000 cycles will be performed. The first two cycles will be performed using the deepest descent method, which is more accurate but takes more time. The remaining 998 cycles will be performed using the conjugate gradient method, which is an approximation of the deepest descent.
The parameter “ntx = 1” indicates that only coordinates but not velocities will be read from the previous step (to read both coordinates and velocities, set it at ntx = 5); “ntc = 1” indicates that the SHAKE algorithm is turned off so that no bonds are constrained (ntc = 2 will constrain any bonds involving H atoms while ntc = 3 will constrain all bonds); “ntb =1” means that a period boundary will be set around the system to maintain a constant volume; “ntp” = 0 means constant pressure will NOT be applied.
The parameters “ntwx”, “ntwe” and “ntpr” define the frequencies at which the program records data during the minimization process: “ntwx = 1000” indicates that coordinates of atoms are deposited into a .trj file every 1000 cycles; “ntwe = 0” indicates that no .en file is being generated; “ntpr = 1000” indicates that energy readings are written into .out and .info files every 1000 steps.
The parameter “scee = 1.2” indicates that we will use a scaling factor of 1.2 for close range 1-4 electrostatic interactions. A cut-off of 8.0 is applied for non-bonded interactions as indicated by “cut = 8.0”.
The flag “ntr =1” indicates that the positional restraint method is applied for the energy minimization (alternatively, something called “BELLY” could be used but we will not to go into any details about that in this tutorial); “restraintmask = ':1-119 & !@H='” indicates that the position of every atom within residues 1 -119 that is NOT a hydrogen is being restrained; “restraint_wt = 5.0” defines how strong the restrain is.
Equilibration
Bear in mind that an energy minimization is fundamentally different from a molecular dynamics simulation because none of the atoms has any velocities associated with it. Essentially, it mimics a state where temperature = 0 K. Obviously this is nothing like what we commonly observe in reality. To prepare the system for the production runs, we will need to heat the system up to 300 K so that it can further equilibrate. To do that, a series of short MD simulations (50,000 frames) with high time resolution (1 fs) with be performed in combination with multiple minimization runs. Listed below is a template input file “10md.in” for MD simulations:
10md.in: production (500000 = 1ns) &cntrl imin = 0, ntx = 5, irest = 1, nstlim = 500000, temp0 = 298.15, tempi = 298.15, ig = 71287, ntc = 2, ntf = 1, ntt = 1, dt = 0.002, ntb = 2, ntp = 1, tautp = 1.0, taup = 1.0, ntwx = 500, ntwe = 0, ntwr = 500, ntpr = 500, scee = 1.2, cut = 8.0, iwrap = 1, ntr = 1, nscm = 100, restraintmask = ':1-118@CA,C,N', restraint_wt = 0.1, /
The flag “imin = 0” indicates that this is an MD simulation instead of an energy minimization; “ntx = 5” indicates that at each step, both coordinates and velocities are inherited from the previous one (as you would expect for an MD simulation); “irest = 1” indicates that everything will restart at the beginning of this run, using information provided in the .rst file generated by the last run; “nstlim = 500000” define the number of steps; “temp0” and “tempi” define the temperature of the system, which is kept at 298.15 throughout all MDs in this case; “ig” defines a random seed for the random number generator. You should pick a different number for “ig” at each restart or simply set it at -1 if you are using AMBER 10 or 11. For the sake of reproducibility we will use the same number for all MD runs.
The flag “ntc =2” indicates that all bonds involving H bonds are constrained by the SHAKE algorithm to eliminate high frequency oscillations in the system; “ntf =1” indicates that all types of forces in the force filed are being calculated; “ntt =1” indicates that the default Berendsen temperature coupling algorithm is being used; “dt” define the length of each frame, which is set at 2 fs here.
The flags “ntb =2” and “ntp =1” indicate that constant pressure instead of constant volume is applied; “taup = 1.0” defines the temperature relaxation time.
Again, “ntwx”, “ntwe”, “ntwr” and “ntpr” define the frequency of data deposition. Note that “ntwr” defines how often the .rst7 file is saved. A high frequency for “ntwr” makes it less painful to recover the system from a crash.
The flag “iwrap = 1” indicate that the coordinates written to the .rst7 and .trj files will be wrapped into a primary box. This reduces the amount of coordinate output, thus preventing overflowing and making visualization more convenient.
The parameter “nscm = 100” indicate that the translational movements of the center-of-mass are removed every 100 frames. For non-periodic systems, rotational movements will also be removed.
Beware that the parameter specified in 10md.in is for a production run (as indicated in the first line). For the minimization and equilibration runs (01 ~ 09), the parameters that differ from the templates “1mi.in” and “10md.in” are listed in the section below:
01mi.in: maxcyc = 1000, restraintmask = ':1-119 & !@H=', restraint_wt = 5.0,
02md.in: ntx = 1, irest = 0, nstlim = 50000, restraintmask = ':1-119 & !@H=',
restraint_wt = 5.0, dt = 0.001,
03mi.in: maxcyc = 1000, restraintmask = ':1-119 & !@H=', restraint_wt = 2.0,
04mi.in: maxcyc = 1000, restraintmask = ':1-119 & !@H=', restraint_wt = 0.1,
05mi.in: maxcyc = 1000, restraintmask = ':1-119 & !@H=', restraint_wt = 0.05,
06md.in: ntx = 1, irest = 0, nstlim = 50000, restraintmask = ':1-119 & !@H=',
restraint_wt = 1.0, dt = 0.001,
ntwx = 1000, ntwr = 1000, ntpr = 1000,
07md.in: nstlim = 50000, restraintmask = ':1-119 & !@H=', restraint_wt = 0.5, dt = 0.001,
ntwx = 1000, ntwr = 1000, ntpr = 1000,
08md.in: nstlim = 50000, restraintmask = ':1-118@CA,C,N', restraint_wt = 0.1, dt = 0.001,
ntwx = 1000, ntwr = 1000, ntpr = 1000,
09md.in: nstlim = 50000, restraintmask = ':1-118@CA,C,N', restraint_wt = 0.1, dt = 0.001,
ntwx = 1000, ntwr = 1000, ntpr = 1000,
As you might have noticed from the parameters shown above, through minimization runs 01, 03, 04 and 05, progressively weaker restraints were applied on the non-H atoms. Similarly, through equilibration runs 02, 05, 06 and 07, restraints also weaker run by run, and a time resolution of 1 femto-second is being used in contrast to 2 femto-seconds in the production runs. In runs 08 and 09, side chains are no longer restrained. Only the heavy atoms of the backbone chain (CA, C and N) are restrained, which makes them essentially the same as the production runs except for the shorter durations (50,000 frames in contrast to 500,000 frames).
Bear in mind that these minimization and MD runs will be performed one after another, followed by the production runs (10 and 11). Although you can manually submit them one by one, it is more convenient and time-saving to write up a script carrying out all the jobs via an MPI. The script will be shown in the next section after we have prepared the input files for the last two (production) runs.
Production
Although we run minimization and production in the same script, they represent distinct stages.
We should run the production phase of the simulation using the same conditions as the final phase of equilibration to prevent an abrupt jump in the potential energy due to a change in simulation conditions.
First create the file 10md.in:
10md.in: production (500000 = 1ns)
&cntrl
imin = 0, ntx = 5, irest = 1, nstlim = 500000,
temp0 = 298.15, tempi = 298.15, ig = 71287,
ntc = 2, ntf = 1, ntt = 1, dt = 0.002,
ntb = 2, ntp = 1, tautp = 1.0, taup = 1.0,
ntwx = 500, ntwe = 0, ntwr = 500, ntpr = 500,
scee = 1.2, cut = 8.0, iwrap = 1,
ntr = 1, nscm = 100,
restraintmask = ':1-118@CA,C,N', restraint_wt = 0.1,
/
Then create similar files with the modified parameters:
For equilibration (same ones as shown in the last section):
06md.in: ntx = 1, irest = 0, nstlim = 50000, restraintmask = ':1-119 & !@H=',
restraint_wt = 1.0, dt = 0.001,
ntwx = 1000, ntwr = 1000, ntpr = 1000,
07md.in: nstlim = 50000, restraintmask = ':1-119 & !@H=', restraint_wt = 0.5, dt = 0.001,
ntwx = 1000, ntwr = 1000, ntpr = 1000,
08md.in: nstlim = 50000, restraintmask = ':1-118@CA,C,N', restraint_wt = 0.1, dt = 0.001,
ntwx = 1000, ntwr = 1000, ntpr = 1000,
09md.in: nstlim = 50000, restraintmask = ':1-118@CA,C,N', restraint_wt = 0.1, dt = 0.001,
ntwx = 1000, ntwr = 1000, ntpr = 1000,
For production:
10md.in: nstlim = 500000, restraintmask = ':1-118@CA,C,N', restraint_wt = 0.1, dt = 0.002, 11md.in: nstlim = 500000, restraintmask = ':1-118@CA,C,N', restraint_wt = 0.1, dt = 0.002,
Here the 10md.in and 11md.in use the pre-equilibrated system.
In The above code we see that we are running for 50000 steps (nstlim = 50000), with a time step of dt where dt of 0.001 is 1fs. We are starting the run with no previous velocity information ntx=1, irest=0. We will print energy output every 500 steps (ntwr=500) and save coordinates every 500 (ntwx=500). We want the initial temperature to be 298.15K (tempi=298.15) and the reference temperature to be 300K (temp0=298.15). Initially, we restraint everything except hydrogens with a restraint weight of 1.0, and with later runs we decrease the restraint weight and only the backbone is restrained. With each future run, we are using velocity information from the previous restart file (ntx=5, irest=1), and switched on constant pressure (ntb=2) with isotropic position scaling (ntp=1).
We should keep in mind that each simulation stage is distinct, so if a simulation terminates prematurely, we can easily continue from the last completed stage using the -c flag. (e.g.,-c 10md.rst7).
To run equilibration and production, Write the tcsh file equil.produc.qsub.csh:
#! /bin/tcsh
#PBS -l nodes=4:ppn=2
#PBS -l walltime=720:00:00
#PBS -o zzz.qsub.out
#PBS -e zzz.qsub.err
#PBS -V
set workdir = "~/AMBER_Tutorial/003.SANDER"
cd ${workdir}
cat $PBS_NODEFILE | sort | uniq
mpirun -n 8 sander.MPI -O -i 01mi.in -o 01mi.out -p ../002.TLEAP/1df8.com.wat.leap.prm7 \
-c ../002.TLEAP/1df8.com.wat.leap.rst7 -ref ../002.TLEAP/1df8.com.wat.leap.rst7 \
-x 01mi.trj -inf 01mi.info -r 01mi.rst7
mpirun -n 8 sander.MPI -O -i 02md.in -o 02md.out -p ../002.TLEAP/1df8.com.wat.leap.prm7 \
-c 01mi.rst7 -ref 01mi.rst7 -x 02md.trj -inf 02md.info -r 02md.rst7
mpirun -n 8 sander.MPI -O -i 03mi.in -o 03mi.out -p ../002.TLEAP/1df8.com.wat.leap.prm7 \
-c 02md.rst7 -ref 02md.rst7 -x 03mi.trj -inf 03mi.info -r 03mi.rst7
mpirun -n 8 sander.MPI -O -i 04mi.in -o 04mi.out -p ../002.TLEAP/1df8.com.wat.leap.prm7 \
-c 03mi.rst7 -ref 03mi.rst7 -x 04mi.trj -inf 04mi.info -r 04mi.rst7
mpirun -n 8 sander.MPI -O -i 05mi.in -o 05mi.out -p ../002.TLEAP/1df8.com.wat.leap.prm7 \
-c 04mi.rst7 -ref 04mi.rst7 -x 05mi.trj -inf 05mi.info -r 05mi.rst7
mpirun -n 8 sander.MPI -O -i 06md.in -o 06md.out -p ../002.TLEAP/1df8.com.wat.leap.prm7 \
-c 05mi.rst7 -ref 05mi.rst7 -x 06md.trj -inf 06md.info -r 06md.rst7
mpirun -n 8 sander.MPI -O -i 07md.in -o 07md.out -p ../002.TLEAP/1df8.com.wat.leap.prm7 \
-c 06md.rst7 -ref 05mi.rst7 -x 07md.trj -inf 07md.info -r 07md.rst7
mpirun -n 8 sander.MPI -O -i 08md.in -o 08md.out -p ../002.TLEAP/1df8.com.wat.leap.prm7 \
-c 07md.rst7 -ref 05mi.rst7 -x 08md.trj -inf 08md.info -r 08md.rst7
mpirun -n 8 sander.MPI -O -i 09md.in -o 09md.out -p ../002.TLEAP/1df8.com.wat.leap.prm7 \
-c 08md.rst7 -ref 05mi.rst7 -x 09md.trj -inf 09md.info -r 09md.rst7
mpirun -n 8 sander.MPI -O -i 10md.in -o 10md.out -p ../002.TLEAP/1df8.com.wat.leap.prm7 \
-c 09md.rst7 -ref 05mi.rst7 -x 10md.trj -inf 10md.info -r 10md.rst7
mpirun -n 8 sander.MPI -O -i 11md.in -o 11md.out -p ../002.TLEAP/1df8.com.wat.leap.prm7 \
-c 10md.rst7 -ref 05mi.rst7 -x 11md.trj -inf 11md.info -r 11md.rst7
exit
Note in the above script that for each run, the .rst7, .trj and .info files generated from the previous run provides the initial state to start from. The .prm7 file generated for the hydrated complex by TLEAP provides the force parameters.
Submit the file to the que and monitor progress.
IV. Simulation Analysis
Ptraj
To run the Ptraj analysis, you will probably first create a separate folder for it under the root directory if you don't have one.
mkdir 004.PTRAJ
We will run five Ptraj analysis so that we need five separate input files. We will first create ptraj.1.in containing the following lines.
trajin ../003.SANDER/10md.trj 1 1000 1 trajin ../003.SANDER/11md.trj 1 1000 1 trajout 1DF8.trj.strip nobox strip :WAT
This will concatenate your two 1ns trajectories together, strip off the waters, and output it as a new file called 1DF8.trj.strip. The two sets of numbers 1 1000 1 give you the input information about which frames you are using for the Ptraj. The first two numbers 1 and 1000 specify the starting and ending snapshots from your trajectory file. The ending number of the snapshot doesn't need to be accurate because if you actually don't have enough snapshots in your trajectory file, Ptraj will read up to the last one you have. The last number 1 specifies the frequency of the snapshot saved, in this case, we are saving every frame of the trajectory file. And the last line of the input file will take away all the water molecules.
To execute the first Ptraj analysis, run the following command specifying both input and out put files.
ptraj ../002.ANTE.TLEAP/1DF8.com.wat.leap.parm ptraj.1.in > ptraj.1.log
After the first step, we now have the 1DF8.trj.strip file which contains 2000 snapshots of the trajectory, and we will compare it to our reference file 1DF8.com.gas.leap.rst7, using the following input file named as ptraj.2.in.
trajin 1DF8.trj.strip 1 2000 1 trajout 1DF8.com.trj.stripfit reference ../002.ANTE.TLEAP/1DF8.com.gas.leap.rst7 rms reference out 1DF8.rmsd.CA.txt :1-118@CA
Note that in this input file, the set of numbers for trajin is 1 2000 1 because in 1DF8.trj.strip we actually have 2000 snapshots. The third line specifies the reference file we are using as 1DF8.com.gas.leap.rst7, because we have taken away all the water molecules in the first step, we are comparing our trajectory to the gas phase complex. And the last line says we are calculating the rmsd for alpha carbon number 1 to 118.
To execute the the analysis, run the following line.
ptraj ../002.ANTE.TLEAP/1DF8.com.gas.leap.parm ptraj.2.in > ptraj.2.log
This will give us an txt file with two columns in it, the first column is the number of frame it is comparing and the second column is the rmsd. And then we will generate a similar rmsd file for ligand as well using the input file as follows.
trajin 1DF8.com.trj.stripfit 1 2000 1 reference ../002.ANTE.TLEAP/1DF8.com.gas.leap.rst7 rms reference out 1DF8.lig.rmsd.txt :119@C*,N*,O*,S* nofit
Similarly, this time we will compare the trajectory file to 1DF8.com.gas.leap.rst7 as well, only this time we are comparing the ligand instead of the receptor by specifying that in the last line saying we want to calculate the rmsd for carbon, nitrogen, oxygen and sulfur in residue 119. And also the <nofit> in the last line specifying that we are calculating the rmsd of the ligand to the crystal structure without any fitting.
To execute the the analysis, run the following line.
ptraj ../002.ANTE.TLEAP/1DF8.com.gas.leap.parm ptraj.3.in > ptraj.3.log
Finally, you want to perform MMGBSA analysis to obtain energetic information about the system. To do this, you need a trajectory file of the gas phase complex (which we already created above and saved as 1DF8.com.trj.stripfit), and want to create two more trajectory files containing just the receptor and just the ligand. To do so, you have to run two more ptraj using the following ptraj input files:
ptraj.4.in (receptor only)
trajin 1DF8.com.trj.stripfit 1 2000 1 trajout 1DF8.rec.trj.stripfit strip :119
ptraj.5.in (ligand only)
trajin 1DF8.com.trj.stripfit 1 2000 1 trajout 1DF8.lig.trj.stripfit strip :1-118
To execute, type in the following commands:
ptraj ../002.ANTE.TLEAP/1DF8.com.gas.leap.parm ptraj.4.in > ptraj.4.log ptraj ../002.ANTE.TLEAP/1DF8.com.gas.leap.parm ptraj.5.in > ptraj.5.log
To visualize trajectories, we have to move the trajectory files to Herbie with scp or rsync commands and run VMD.
rsync -arv 004.PTRAJ/ ID@compute.mathlab.sunysb.edu:~AMS536/AMBER_tutorial/004.PTRAJ
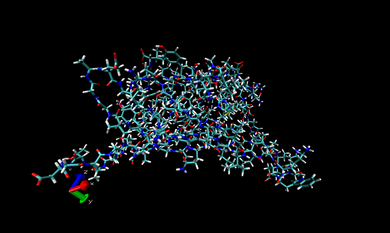
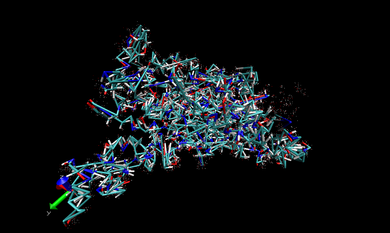
In VMD, we open one of the .prm7 files in 002.ANTE.TLEAP. If you want to visualize the whole complex in gas state, you can open 1DF8.com.gas.leap.prm7 with AMBER7 Parm from 002.ANTE.TLEAP and then 1DF8.com.trj.stripfit with AMBER coordinates from 004.PTRAJ. With these files, you can look at the real-time movement of the complex in the gas state like following snapshot.
You can repeat this procedure to observe the real-time movement of the complex in the water state. Only different thing is opening 1DF8.com.wat.leap.prm7 instead of 1DF8.com.gas.leap.prm7. The following snapshot is that of movement in the water phase.
In the snapshot above, the water molecules are shown as points.
RMSD Plots
Once you have finished running Ptraj, you should have two RMSD files: 1df8.lig.rmsd.txt 1df8.rmsd.CA.txt
The data in these files can be plotted to visualize the structural drift (ie change in RMSD) over the course of your simulation. Large fluctuations in RMSD may be an indication that your simulation is unstable.
Here is an example plot for ligand RMSD:
Measuring h-bond distances
Ptraj can also be used to measure the hydrogen bonding distance between two atoms. To use this functionality, we first need to decide which atom pairs are forming interesting h-bonds. We can do this with VMD, which can draw H-bonds automatically for us. First, select the ligand (BTN) and residues around the ligand. Second, change the Drawing Method to HBonds.
After you have picked out some atom pairs, use the distance command in Ptraj to measure the distance between the two atoms.
Here is a sample script:
#! /bin/tcsh
#PBS -l nodes=1:ppn=1
#PBS -l walltime=01:00:00
#PBS -o zzz.qsub.out
#PBS -e zzz.qsub.err
#PBS -V
set workdir = "~/AMBER_tutorial/007.PTRAJ.HBONDS"
cd ${workdir}
ptraj ../002.TLEAP/1df8.com.gas.leap.prm7 << EOF
trajin ../004.PTRAJ/1df8.com.trj.stripfit
distance 34N_119O2 :34@N :119@O2 out 34N_119O2.out
distance 73OG_119O3 :73@OG :119@O3 out 73OG_119O3.out
distance 12OG_119O14 :12@OG :119@O14 out 12OG_119O14.out
distance 28OH_119O14 :28@OH :119@O14 out 28OH_119O14.out
EOF
The results can be easily plotted by writing a script for XMGRACE. Here is an example:
#!/bin/tcsh foreach distance (28OH_119O14 12OG_119O14) cat << EOF > $distance.grace.in READ NXY "$distance.out" s0 legend "$distance" title "Example Hydrogen Bond Distance Plot" xaxis label "Time (picoseconds)" yaxis label "Hydrogen Bond Distance (Angstroms)" PRINT TO "$distance.eps" DEVICE "EPS" OP "level2" PRINT EOF xmgrace -batch $distance.grace.in -nosafe -hardcopy end
This will generate two plots for the above hydrogen bond distances in EPS format, here is one of them:
MMGBSA Energy Calculation
MM/GBSA is the acronym for Molecular Mechanics/Generalized Born Surface Area. This part of AMBER combines molecular mechanics (MM) with both the electrostatic (PB) and nonpolar (SA) contribution to solvation . Topology files are needed for the receptor (R), ligand (L), and receptor-ligand complex (RL). The trajectory files (trj) generate the coordinates. Therefore, molecular dynamics is used to generate a set of snapshots taken at fixed intervals from the trajectories. These snapshots are processed to remove solvent and generate the free energy of binding.
In the AMBER_Tutorial directory, create a new directory:
mkdir 005.MMGBSA
In this new directory, create the file gb.rescore.in containing:
Single point GB energy calc &cntrl ntf = 1, ntb = 0, ntc = 2, idecomp= 0, igb = 5, saltcon= 0.00, gbsa = 2, surften= 1.0, offset = 0.09, extdiel= 78.5, cut = 99999.0, nsnb = 99999, scnb = 2.0, scee = 1.2, imin = 5, maxcyc = 1, ncyc = 0, /
Then create a csh script, run.sander.rescore.csh, that contains the following lines of command:
#! /bin/tcsh
#PBS -l nodes=1:ppn=2
#PBS -l walltime=24:00:00
#PBS -o zzz.qsub.out
#PBS -e zzz.qsub.err
#PBS -V
set workdir = “${HOME}/AMBER_Tutorial/005.MMGBSA”
cd ${workdir}
sander -O -i gb.rescore.in \
-o gb.rescore.out.com \
-p ../002.TLEAP/1df8.com.gas.leap.prm7 \
-c ../002.TLEAP/1df8.com.gas.leap.rst7 \
-y ../004.PTRAJ/1df8.com.trj.stripfit \
-r restrt.com \
-ref ../002.TLEAP/1df8.com.gas.leap.rst7 \
-x mdcrd.com \
-inf mdinfo.com
sander -O -i gb.rescore.in \
-o gb.rescore.out.lig \
-p ../002.TLEAP/1df8.lig.gas.leap.prm7 \
-c ../002.TLEAP/1df8.lig.gas.leap.rst7 \
-y ../004.PTRAJ/1df8.lig.trj.stripfit \
-r restrt.lig \
-ref ../002.TLEAP/1df8.lig.gas.leap.rst7 \
-x mdcrd.lig \
-inf mdinfo.lig
sander -O -i gb.rescore.in \
-o gb.rescore.out.rec \
-p ../002.TLEAP/1df8.rec.gas.leap.prm7 \
-c ../002.TLEAP/1df8.rec.gas.leap.rst7 \
-y ../004.PTRAJ/1df8.rec.trj.stripfit \
-r restrt.rec \
-ref ../002.TLEAP/1df8.rec.gas.leap.rst7 \
-x mdcrd.rec \
-inf mdinfo.rec
exit
Then this script should be sent to the queue, i.e., qsub the script using the commands:
chmod +x run.sander.rescore.csh qsub run.sander.rescore.csh
You can monitor your progress by typing qstat -u username.
When the job is complete, you will obtain the following output files: gb.rescore.out.com, gb.rescore.out.lig, and gb.rescore.out.rec
In these files, the single point energy calculation results will be written for each individual frame. It will be found in the results section and the output file will have an infrastrucutre that is similar to the following:
FINAL RESULTS
NSTEP ENERGY RMS GMAX NAME NUMBER
1 7.9662E+03 1.9504E+01 1.3272E+02 C 3403
BOND = 836.7674 ANGLE = 2343.1208 DIHED = 2917.9356
VDWAALS = -2299.0570 EEL = -19265.6215 EGB = -3539.4050
1-4 VDW = 1071.1234 1-4 EEL = 11631.2887 RESTRAINT = 0.0000
ESURF = 14270.0746
minimization completed, ENE= 0.79662270E+04 RMS= 0.195036E+02
minimizing coord set # 2
The results generated in the output files will be used for the final calculations. Use the grep command for complex (com), ligand (lig), and receptor (rec) to obtain the results.
In the command line, type:
grep VDWAALS gb.rescore.out.com > vdw.com.txt.
grep ESURF gb.rescore.out.com > surf.com.txt.
You can take these text files, import them into Excel, and do the rest of your modifications there.
Equations for analysis
Remember that to obtain the Gvdw term, you need to take the SASA (which is ESURF) and input into equation 1:
ΔGnonpolar = SASA*0.00542 + 0.92 [1]
Also, the mmgbsa of a given system can be determined by equation 2:
ΔGmmgbsa = ΔGvdw + ΔGcoul + ΔGpolar + ΔGnonpolar [2]
From the output file:
VDWAALS = ΔGvdw
EELS = ΔGcoul
EGB = ΔGpolar
You can then easily calculate the ΔΔGbind by using equation 3:
ΔΔGbind = ΔGmmgbsa,complex – (ΔGmmgbsa,lig + ΔGmmgbsa,rec) [3]
You will want to careful when doing your analysis that the results from frame 1 for the receptor and ligand are subtracted from the results from frame 1 for your complex. By doing this in excel, you should have 2000 frames for each, and the values should cleanly line up. Finally, you will want to plot your ΔΔGbind and examine if you see corresponding changes in the ligand position and the ΔΔGbind. Also, you should determine the mean and standard deviation for your ΔΔGbind.
Plotting your results
When your rescoring calculation finishes, you have the following three output files: "gb.rescore.out.com", "gb.rescore.out.lig", and "gb.rescore.out.rec".
Use the following script, entitled get.mmgbsa.bash, to extract data and calculate MMGBSA energy for each snap shot.
#! /bin/bash
# by Haoquan
echo com lig rec > namelist
LIST=`cat namelist`
for i in $LIST ; do
grep VDWAALS gb.rescore.out.$i | awk '{print $3}' > $i.vdw
grep EGB gb.rescore.out.$i | awk '{print $9}' > $i.polar
grep EELS gb.rescore.out.$i | awk '{print $6}' > $i.coul
grep ESURF gb.rescore.out.$i | awk '{print $3 * 0.00542 + 0.92}' > $i.surf
paste -d " " $i.vdw $i.polar $i.surf $i.coul | awk '{print $1 + $2 + $3 + $4}' > data.$i
rm $i.*
done
paste -d " " data.com data.lig data.rec | awk '{print $1 - $2 - $3}' > data.all
for ((j=1; j<=`wc -l data.all | awk '{print $1}'`; j+=1)) do
echo $j , >> time
done
paste -d " " time data.all > MMGBSA_vs_time.dat
rm namelist time data.*
To run this script do:
chmod +x get.mmgbsa.sh
bash get.mmgbsa.sh
This will create a text file called MMGBSA_vs_time.dat with x and y values separated by a space and comma. These values can be imported to Excel or Origin or to XMGRACE if you are using Linux:
xmgrace MMGBSA_vs_time.dat
V. Frequently Encountered Problems
Barbara
1.If you have a problem opening the structure of your molecule using VMD, be certain that the prm7 file corresponds to the rst7 file. For example, 1df8.lig.wat.leap.prm7 must be matched to 1df8.lig.wat.leap.rst7.
2. The VMD Console window (often hidden behind the visualization screen of your molecule) can be used to determine where VMD has found an error in your typed commands if you are unable to visualize your molecule.
3. If your "rescore" files do not appear when you are running the script: run.sander.rescore.csh, it may be the result of there not being a free node. You job may be sitting on the queue. If you type qstat -u username, you will be able to see if the job is sitting there because a Q will appear.
Woo Suk
How to run AMBER with DNA molecule
If you have a DNA molecule as a substrate, you cannot run antechamber to add hydrogen atoms and charge. The antechamber is only for small molecules. In this case, you don't have to save ligand file as .mol2. Save the ligand file with pdb format and just run tleap like protein. Only different thing compared to protein tleap.in file is the force field. For DNA molecules, you have to use ff99bsc0 forcefield instead of ff99sb like following:
set default PBradii mbondi2 source leaprc.ff99bsc0 loadoff rizzo_amber7.ionparms/ions.lib loadamberparams rizzo_amber7.ionparms/ions.frcmod lig = loadpdb ../001.CHIMERA.MOL.PREP/3tmm.lig.pdb saveamberparm lig 3tmm.lig.gas.leap.prm7 3tmm.lig.gas.leap.rst7 solvateBox lig TIP3PBOX 10.0 saveamberparm lig 3tmm.lig.wat.leap.prm7 3tmm.lig.wat.leap.rst7 charge lig quit
In DNA pdb file, you have to note that the atom names must be DA, DT, DG and DC format. Your parameter file cannot recognize other name format like AD or dA.
Longfei
When you are running ptraj, make sure that you read the output .log file each time, because this file will contain the error message when you make a mistake. It is quite common that you make a mistake during simulation, but the important thing is that you know how to find the errors and fix the problems.
Roman
One of the possible problems with running the production step of Sander is that the computers being utilized lose power, or the run is otherwise compromised. In such cases there are two options to finish the simulation. One easy way is to delete all progress at the stage that was interrupted and continue anew from the previous stage. For example, if we lose progress at stage 11, we would just rerun this section:
mpirun -n 8 sander.MPI -O -i 11md.in -o 11md.out -p ../002.TLEAP/1df8.com.wat.leap.prm7 \ -c 10md.rst7 -ref 05mi.rst7 -x 11md.trj -inf 11md.info -r 11md.rst7 exit
This would continue on from stage 10.
Additionally, one can continue from the step that got interrupted using information from *.out file. For example, a line like this:
10md.out: NSTEP = 100500 TIME(PS) = 401.000 TEMP(K) = 298.39 PRESS = -123.4
in 10md.out indicates that the system reached at least step 100500. So one can continue from that step in the next stage.
Quan
Hui
Tuoling
Some important things when modeling protein-protein interaction in AMBER
In structural preparation step, you should treat both proteins as receptors. Remove the water molecules and other small molecules needed. (Do not add hydrogens and charges in this case). Save the file into pdb fomat as Woo Suk wrote. You may need to modify the some pdb files. E.g. change residue name CYS to CYX, change SH to SG otherwise it will not be recognized by tleap.
Plot per-residue RMSF (Root mean square fluctuation) value to 3D structure
Generate RMSF values using ptraj: creat ptraj.in in vim. Write scripts like this.
trajin 1G9M.com.trj.stripfit 1 2000 1 reference ../002.ANTE.TLEAP/1G9M.com.gas.leap.rst7 average avg.pdb pdb atomicfluct out RMSF.dat :1-486@CA byres
run ptraj:
ptraj ../002.ANTE.TLEAP/1G9M.com.gas.leap.prm7 ptraj.in > ptraj.log
you will get two output files: avg.pdb and RMSF.dat. You can generate B factor data instead of RMSF by adding "bfactor" to the end of last line. To calculate any other properties, you can refer to the AMBER Tools User's Manual.
Map RMSF to 3D structure:
copy the second column of RMSF.dat file to the last column (beta column) of avg.pdb. You can make a "perl" script, or use excel to do that. In my case, I type "vimdiff avg.pdb RMSF.dat" to open two files together. Then use visual block (ctrl + v) to copy (y) and paste (p).
You can copy any column of per-residue properties to the beta column of pdb file you care about. E.g. RMSD, energy, footprint interactions, sequence conservation.
Then you can use vmd to draw the plot: VMD Main --> Graphics --> Representation --> Coloring Method --> Beta. You can change the color scale: VMD Main --> Graphics --> color --> Color Scale --> Method --> whatever color range you like.
Yuchen
One important thing is that you should always make sure that you have the right ligand, receptor and complex structure before you run Ptraj. Because Ptraj is a relatively expensive calculation, for example in this tutorial, the calculation takes around 80 hours when using 8 processors for parallel computing, you don't want to find out that your results are not right simply because you have a typo in your TLEAP input file. What you can do is that after you generate the parameter and restart file using TLEAP, you can use VMD to visually check if you have the right structure.
Yunting
Before the MD simulation, we should check the receptor, ligand and complex structure files to make sure they are accurate and compatible with each other. We can check the files directly or use some visualization tool like VMD to do this.
Kip
PTRAJ Script
PTRAJ can only be run in serial mode (ie, only on one single processor) but it can still be useful to make a script and submit your job to Seawulf. Here is an example an example script that will run all 5 PTRAJ jobs described above:
#! /bin/tcsh
#PBS -l nodes=1:ppn=1
#PBS -l walltime=01:00:00
#PBS -o zzz.qsub.out
#PBS -e zzz.qsub.err
#PBS -V
set workdir = "~/AMBER_tutorial/004.PTRAJ"
cd ${workdir}
ptraj ../002.TLEAP/1df8.com.wat.leap.prm7 ptraj.1.in > ptraj.1.log
ptraj ../002.TLEAP/1df8.com.gas.leap.prm7 ptraj.2.in > ptraj.2.log
ptraj ../002.TLEAP/1df8.com.gas.leap.prm7 ptraj.3.in > ptraj.3.log
ptraj ../002.TLEAP/1df8.com.gas.leap.prm7 ptraj.4.in > ptraj.4.log
ptraj ../002.TLEAP/1df8.com.gas.leap.prm7 ptraj.5.in > ptraj.5.log
exit
.crd vs. .rst7 files
Note that .crd files are the same as .rst7 -- you can simply change the extension if you wish.
Simulation with explicit solvent
Note that if you have (explicit) waters in your AMBER simulation, you will need to use a periodic box
XMGRACE on Mac OS
You'll need to install the OpenMotif libraries first: http://www.ist.co.uk/downloads/motif_download.html
Then do:
cd Desktop wget ftp://ftp.fu-berlin.de/unix/graphics/grace/src/grace5/grace-latest.tar.gz tar xzvf grace-latest.tar.gz cd grace-5.1.22 ./configure --x-includes=/usr/X11/include --x-libraries=/usr/X11/lib \ --with-motif-library=-lXm --with-extra-incpath=/usr/OpenMotif/include:/Users/sm4082/fftw-2.1.5/include \ --with-extra-ldpath=/usr/OpenMotif/lib:/Users/sm4082/fftw-2.1.5/lib make sudo make install echo "export PATH=$PATH:/usr/local/grace/bin" >> ~/.bashrc source ~/.bashrc xmgrace
Alternatively, you can also install XMGRACE using Fink or MacPorts
Running out of disk space
If you fill up your quota of disk space on Herbie, you can temporarily store files in the scratch space located at /space1
Please note that /space1 is for temporary storage only and this space is periodically erased. Nevertheless, this scratch space is very useful for moving files from Seawulf to Herbie and then to your personal computer for permanent storage (or for temporarily storing files that you want to visualize on a math lab computer, etc.)