Difference between revisions of "2019 Covalent docking tutorial 1 with PDB 5VKG"
Stonybrook (talk | contribs) (→III. Preparing the sphere files) |
|||
| (12 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | This tutorial teaches you how to dock a covalently bound drug molecule to a receptor (PDB 5VKG). | + | This tutorial teaches you how to dock a covalently bound drug molecule to a receptor (PDB 5VKG) using an alpha version of DOCK6.10. |
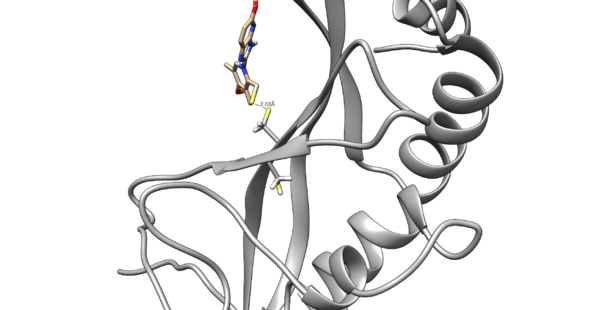
Use chimera to visualize your system in this case the pdb 5VKG of NMR structure of human Tsg101 UEV in complex with tenatoprazole. | Use chimera to visualize your system in this case the pdb 5VKG of NMR structure of human Tsg101 UEV in complex with tenatoprazole. | ||
| − | PDB 5VKG consists of 20 different models, but we need only one. Either chose one of the models or ask Lauren to give you the file rcsb104645_model1.pdb | + | PDB 5VKG consists of 20 different models, but we need only one. Either chose one of the models or ask Lauren to give you the file rcsb104645_model1.pdb. Also ask Lauren to provide you with a necessary version of dock6. |
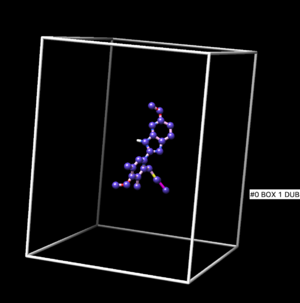
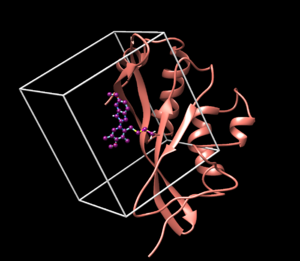
[[Image:image_starting_structure.png|thumb|center|600px| N16 covalently bound to Tsg101 is shown]] | [[Image:image_starting_structure.png|thumb|center|600px| N16 covalently bound to Tsg101 is shown]] | ||
| Line 69: | Line 69: | ||
2 '''D1''' -25.1580 36.1840 -35.7130 '''Du''' 1 LIG -0.1111 | 2 '''D1''' -25.1580 36.1840 -35.7130 '''Du''' 1 LIG -0.1111 | ||
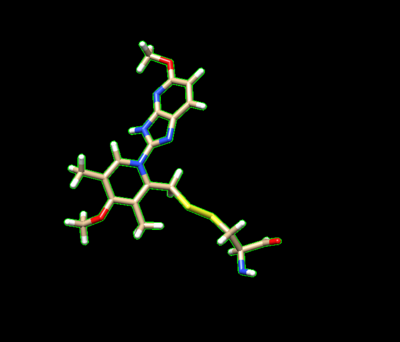
| + | Delete the backbone of the cysteine leaving the beta carbon. | ||
The ligand should now look like this in chimera: | The ligand should now look like this in chimera: | ||
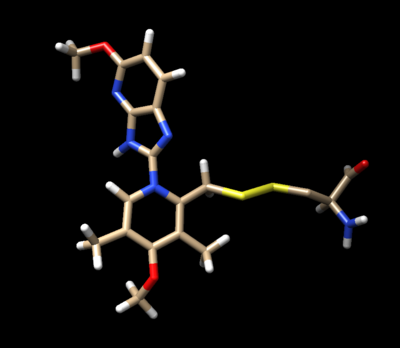
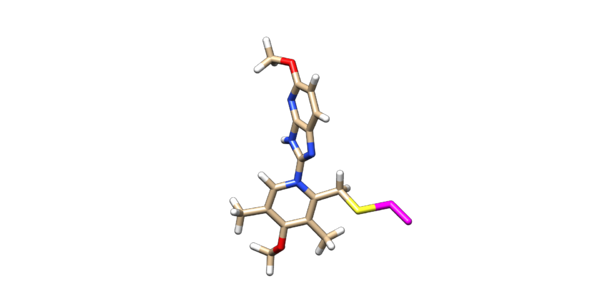
[[Image:Image_n16_charged_with_dummy_atoms.png|thumb|center|600px| N16 modified to include the beta carbon and sulfur as dummy atoms. ]] | [[Image:Image_n16_charged_with_dummy_atoms.png|thumb|center|600px| N16 modified to include the beta carbon and sulfur as dummy atoms. ]] | ||
| Line 81: | Line 82: | ||
[[Image:Image_n16_spheres.png|thumb|center|600px| The three atoms from the cysteine sidechain needed for oritenting: beta carbon, alpha carbon, and sulfur.]] | [[Image:Image_n16_spheres.png|thumb|center|600px| The three atoms from the cysteine sidechain needed for oritenting: beta carbon, alpha carbon, and sulfur.]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | Convert saved pdb or mol2 into a sphere file | + | Convert saved coordinates in pdb or mol2 into a sphere file: lig_abs.mol2 lig_abs_spheres.sph |
| − | |||
[[Image: Ligabsspheres.png |thumb|center|300px| Spheres for Orienting]] | [[Image: Ligabsspheres.png |thumb|center|300px| Spheres for Orienting]] | ||
| Line 153: | Line 96: | ||
| − | Now spheres will be generated for the box. A sphere file will be made using the coordinates from the ligand file lig_03_06.mol2. | + | Now spheres will be generated for the box. A sphere file will be made using the coordinates from the ligand file lig_03_06.mol2. Use only heavy atoms to generate spheres. |
| + | |||
| − | |||
| − | [[Image: | + | [[Image: Ligspheres2.png |thumb|center|300px| Spheres for Orienting]] |
| + | |||
| + | == '''IV. Box generating''' == | ||
| + | |||
| + | Now that the spheres are generated twice once for orientating and the other for the dock/grid. The box will be generated. | ||
| + | Go to 003.gridbox. Create a file showbox.in. In showbox.in write: | ||
| + | Y | ||
| + | 8.0 | ||
| + | ../002.spheres/lig_spheres.sph | ||
| + | 1 | ||
| + | lig.box.pdb | ||
| + | |||
| + | Run the command to generate a box: | ||
| + | showbox < showbox.in | ||
| + | |||
| + | The box result is shown at Figure below. | ||
| + | [[Image:box1.png |thumb|center|300px| Spheres in box. Heavy atoms in ligand's spheres were used to create a box.]] | ||
| + | |||
| + | |||
| + | == '''V. Grid generating''' == | ||
| + | |||
| + | Once the box is made the grid can be populated in the box. | ||
| + | Create a file grid.in. But before that copy vdw.defn and chem.defn in current working directory. If don't do this the grid could not recognize the long path for this parameter. | ||
| + | Write below into the grid.in file: | ||
| + | compute_grids yes | ||
| + | grid_spacing 0.4 | ||
| + | output_molecule no | ||
| + | contact_score no | ||
| + | energy_score yes | ||
| + | energy_cutoff_distance 9999 | ||
| + | atom_model a | ||
| + | attractive_exponent 6 | ||
| + | repulsive_exponent 9 | ||
| + | distance_dielectric yes | ||
| + | dielectric_factor 4 | ||
| + | bump_filter yes | ||
| + | bump_overlap 0.75 | ||
| + | receptor_file ../001.files/5VKG_03_06.mol2 change it to ../001.files/5VKG_rec_h.mol2 | ||
| + | box_file lig.box.pdb | ||
| + | vdw_definition_file vdw.defn | ||
| + | chemical_definition_file chem.defn | ||
| + | score_grid_prefix grid | ||
| + | |||
| + | Run | ||
| + | grid -i grid.in -o grid.out | ||
| + | |||
| + | If this is successful, you will get following output files. | ||
| + | grid.out | ||
| + | grid.nrg | ||
| + | grid.bmp | ||
| + | |||
| + | We can begin covalent docking. | ||
| + | |||
| + | == '''VI. Docking''' == | ||
| + | Before starting doing docking open all the prepared structures in chimera to visually observe that everything is prepared correctly. Although when everything looks fine, it still doesn't mean that we prepared everything correctly. But if something looks definitely weird, for example the receptor and the box with a ligand are far from each other, we should think about it and redo. | ||
| + | |||
| + | [[Image:Lig_rec_box.png|thumb|center|300px| Spheres in box. Heavy atoms in ligand's spheres were used to create a box.]] | ||
| + | |||
| + | We can continue with docking. | ||
| + | Create a file covalent.in and write the following into it. | ||
| + | |||
| + | conformer_search_type covalent | ||
| + | pruning_use_clustering yes | ||
| + | pruning_max_orients 100 | ||
| + | pruning_clustering_cutoff 100 | ||
| + | bondlength 1.8 | ||
| + | dihedral_step 1.0 | ||
| + | pruning_conformer_score_cutoff 100.0 | ||
| + | use_clash_overlap no | ||
| + | write_growth_tree no | ||
| + | use_internal_energy yes | ||
| + | Internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file ../001.files/lig_03_06.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd yes | ||
| + | rmsd_reference_filename ../001.files/lig_03_06.mol2 | ||
| + | use_database_filter no | ||
| + | orient_ligand yes | ||
| + | automated_matching yes | ||
| + | receptor_site_file ../002.spheres/lig_abs_spheres.sph | ||
| + | max_orientations 1000 | ||
| + | critical_points no | ||
| + | chemical_matching no | ||
| + | use_ligand_spheres no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | grid_score_primary yes | ||
| + | grid_score_rep_rad_scale 1 | ||
| + | grid_score_vdw_scale 1 | ||
| + | grid_score_es_scale 1 | ||
| + | grid_score_grid_prefix ../003.gridbox/grid | ||
| + | minimize_ligand yes | ||
| + | minimize_anchor no | ||
| + | minimize_flexible_growth yes | ||
| + | use_advanced_simplex_parameters no | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_grow_max_iterations 0 | ||
| + | simplex_grow_tors_premin_iterations 1000 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min yes | ||
| + | simplex_coefficient_restraint 10.0 | ||
| + | atom_model all | ||
| + | vdw_defn_file ../vdw_AMBER_parm99.defn | ||
| + | flex_defn_file ../flex.defn | ||
| + | flex_drive_file ../flex_drive.tbl | ||
| + | ligand_outfile_prefix covalent_out | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1000 | ||
| + | write_conformations no | ||
| + | cluster_conformations yes | ||
| + | cluster_rmsd_threshold 2.0 | ||
| + | rank_ligands no | ||
| + | |||
| + | Run the docking | ||
| + | dock6 -i covalent.in -o covalent.out | ||
| + | |||
| + | To see the output of docking in chimera | ||
| + | Open Chimera | ||
| + | File -> Open -> 5VKG_rec_h.mol2 (Actions -> Surface -> Show) | ||
| + | File -> Open -> lig_03_06.mol2 | ||
| + | |||
| + | Once everything is loaded go to the ViewDock window and use it's menu to view all the calculated properties regarding the covalent docked ligand by following the steps below. | ||
| + | Tools -> Surface/binding Analysis -> ViewDock -> Select the Covalent Dock output file. (covalent_out_scored.mol2) | ||
| + | In the loaded dialog box select Dock4,5 or 6 | ||
| + | Column -> Show -> Grid_Score | ||
| + | Column -> Show -> HA_RMSDs | ||
| + | |||
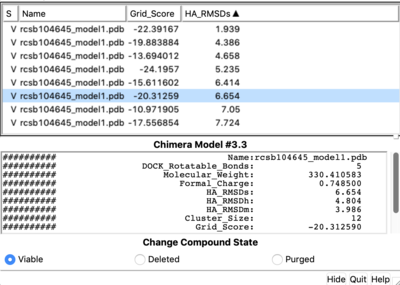
| + | [[Image: Dock_result.png|thumb|center|400px| Dock results]] | ||
| + | |||
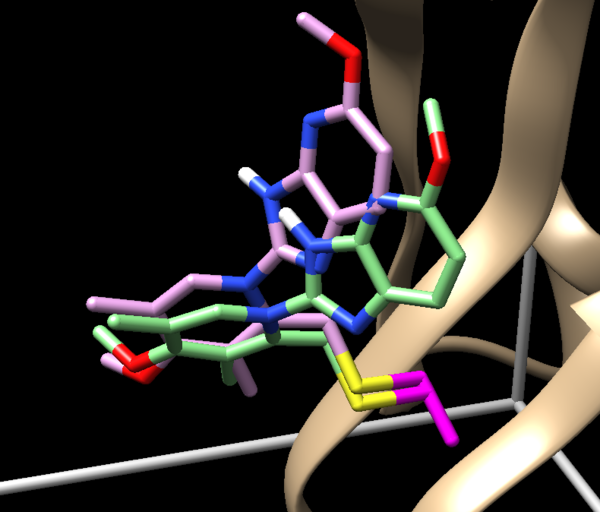
| + | [[Image: Lowest_rmsd_19.png|thumb|center|600px| Docked molecule with the lowest rmsd among all docked poses. Rmsd is 1.9A, -22 Grid score]] | ||
| + | 1.9 RMSD is a sampling success. Is it also a scoring success? To understand it we nee to see which of the molecules was ranked first. TO do this let's open covalent_out_scored.mol2 in text editor and see which ligand is appeared first. If it is a molecule with a lowest RMSD than it is a success, but in our case it is not. It means it is a scoring failure. The lowest RMSD structure is ranked second. | ||
| − | + | [[Image: Dock_first.png|thumb|center|400px| Docked molecule with the best scoring. We see it is a scoring failure as it is not a lowest rmsd ligand pose]] | |
Latest revision as of 16:01, 20 July 2020
This tutorial teaches you how to dock a covalently bound drug molecule to a receptor (PDB 5VKG) using an alpha version of DOCK6.10.
Use chimera to visualize your system in this case the pdb 5VKG of NMR structure of human Tsg101 UEV in complex with tenatoprazole. PDB 5VKG consists of 20 different models, but we need only one. Either chose one of the models or ask Lauren to give you the file rcsb104645_model1.pdb. Also ask Lauren to provide you with a necessary version of dock6.
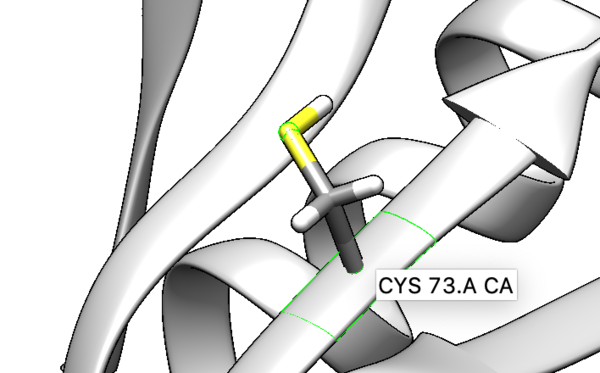
We are interested in the interaction between S atom in Cys 73 and S atom in the ligand. Those two atoms are covalently bonded according to the experimental paper, but Chimera will not show a covalent linker between these two S. We can create the bond ourself in Chimera, if we need (but we don't actually) and measure the distance between these two atoms. If there is a disulfide bond between them, the distance should be 2.05A.
To check the distance we need Shift+Control+left mouse to select two atoms between we want to measure distance.
Tools -> Structure analysis -> Distance.
You will notice the distance from the two sulfurs is about 2.03 angstroms.
Contents
I. Receptor Preparation
First step is to prepare the receptor. To do this open up the original pdb of the molecule. To prepare the receptor, you need to cut off the covalent linkers to generate the receptor.
Tools -> Structure editing -> Build Structure -> Modify structure - Element S, Bonds 2, Geometry tetrahedral.
The newly formed receptor is now with a complete cysteine.
Now add hydrogens and charge the protein. To make the cysteine neutral, protons will be added into the system. Protonate with the latest AMBER force field, currently AMBER14SB.
Tools -> Structure editing -> AddH. Tools -> Structure editing -> AddCharge.
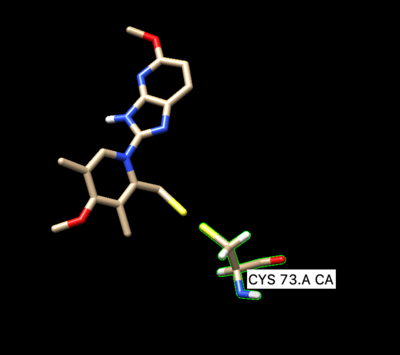
To prepare the receptor for making a grid, delete the side chain of the covalently bonded cysteine residue Cys73. When deleting the side chain of the residue make sure to delete S and the beta carbon, but leave the alpha carbon. That is the receptor that will be used for dock grid and docking.
II. Ligand Preparation
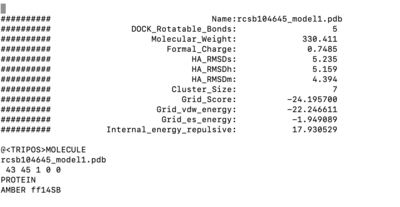
Open of the original structure rcsb104645_model1.pdb. Delete all the protein leaving only the ligand covalently bonded too the cystine, leave the cystine attached to it. The ligand should be binded to the side chain residue all the way up to the beta carbon.
Chose (holding Control + z) Ligand and Cys -> Invert -> Actions -> Atoms -> delete.
Make Cys73 to be visible in all atom mode.
Chose Cys73 (holding Control + z) -> atom/bond -> show.
Then we create a disulfide bond between S in the ligand and the residue. The command bellow will create a disulfide covalent bond:
Chose S in the ligand and in the Cys73 (control +z). Preferences -> Command line -> type "bond sel". It will create a disulfide covalent bond.
Protonate the modified ligand.
Tools -> Structure editing -> AddH
Look at N at the backbone on Cys73 residue. There is only one H attached to N after adding hydrogens, but need to be two. So add one more H to N at the Cys aminoacid backbone. To do so chose N (holding Control +z) and go to
Tools -> Structure editing -> Built structure - Modify Structure - Element N - Bonds 3 - Geometry trigonal.
Next we charge the ligand.
Tools -> Structure editing -> AddCharge -Net charge +1 - with AM1BCC.
Then delete all of the hydrogens off the beta-carbon.
Next we need to modify the beta carbon to D2, and modify the side chain sulfur atom to D1. To do so save prepared ligand in mol2 format and open it with a text editing tool.
Then manually change the atom name of the beta carbon to D2 and change the atom type from C.3 to Du. Change the atom name of the gamma sulfur connected to the beta carbon to D1 and changed the atom type to Du as seen below:
@<TRIPOS>ATOM
1 D2 -24.0727 34.9285 -36.1952 Du 1 LIG -0.0660
2 D1 -25.1580 36.1840 -35.7130 Du 1 LIG -0.1111
Delete the backbone of the cysteine leaving the beta carbon. The ligand should now look like this in chimera:
This is the end of the ligand preparation.
III. Preparing the sphere files
We need to prepare two types of spheres: 1) spheres for orienting the ligand; 2) spheres for box generating .
The only spheres that will be orientated will be the alpha carbon, beta carbon, and the sulfur on the cysteine sidechain. Open the mol2 file with prepared ligand. Isolate the sulfur, alpha and beta carbons and delete everything else. Save it into lig_abs.mol2.
Convert saved coordinates in pdb or mol2 into a sphere file: lig_abs.mol2 lig_abs_spheres.sph
Open file lig_abs_spheres.sph with text editor. Spheres in lig_abs_spheres.sph should be in exact order as 1)S gamma 2)C beta 3)C alpha. Check how atoms are oriented in lig_abs.mol2.
@<TRIPOS>ATOM
1 CA -24.6380 34.0060 -37.3390 C.3 1 CYS 0.0000
2 CB -24.0170 34.8640 -36.2200 C.3 1 CYS 0.0000
3 SG -25.1580 36.1840 -35.7130 S.3 1 CYS 0.0000
It means that in lig_abs_spheres.sph they are also oriented in the same order, but wee need a different order of spheres. Open lig_abs_spheres.sph and rearrange line in correct order by hand.
Now spheres will be generated for the box. A sphere file will be made using the coordinates from the ligand file lig_03_06.mol2. Use only heavy atoms to generate spheres.
IV. Box generating
Now that the spheres are generated twice once for orientating and the other for the dock/grid. The box will be generated. Go to 003.gridbox. Create a file showbox.in. In showbox.in write:
Y 8.0 ../002.spheres/lig_spheres.sph 1 lig.box.pdb
Run the command to generate a box:
showbox < showbox.in
The box result is shown at Figure below.
V. Grid generating
Once the box is made the grid can be populated in the box. Create a file grid.in. But before that copy vdw.defn and chem.defn in current working directory. If don't do this the grid could not recognize the long path for this parameter. Write below into the grid.in file:
compute_grids yes grid_spacing 0.4 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4 bump_filter yes bump_overlap 0.75 receptor_file ../001.files/5VKG_03_06.mol2 change it to ../001.files/5VKG_rec_h.mol2 box_file lig.box.pdb vdw_definition_file vdw.defn chemical_definition_file chem.defn score_grid_prefix grid
Run
grid -i grid.in -o grid.out
If this is successful, you will get following output files.
grid.out grid.nrg grid.bmp
We can begin covalent docking.
VI. Docking
Before starting doing docking open all the prepared structures in chimera to visually observe that everything is prepared correctly. Although when everything looks fine, it still doesn't mean that we prepared everything correctly. But if something looks definitely weird, for example the receptor and the box with a ligand are far from each other, we should think about it and redo.
We can continue with docking. Create a file covalent.in and write the following into it.
conformer_search_type covalent pruning_use_clustering yes pruning_max_orients 100 pruning_clustering_cutoff 100 bondlength 1.8 dihedral_step 1.0 pruning_conformer_score_cutoff 100.0 use_clash_overlap no write_growth_tree no use_internal_energy yes Internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../001.files/lig_03_06.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes rmsd_reference_filename ../001.files/lig_03_06.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.spheres/lig_abs_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid minimize_ligand yes minimize_anchor no minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_grow_max_iterations 0 simplex_grow_tors_premin_iterations 1000 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file ../vdw_AMBER_parm99.defn flex_defn_file ../flex.defn flex_drive_file ../flex_drive.tbl ligand_outfile_prefix covalent_out write_orientations no num_scored_conformers 1000 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
Run the docking
dock6 -i covalent.in -o covalent.out
To see the output of docking in chimera
Open Chimera File -> Open -> 5VKG_rec_h.mol2 (Actions -> Surface -> Show) File -> Open -> lig_03_06.mol2
Once everything is loaded go to the ViewDock window and use it's menu to view all the calculated properties regarding the covalent docked ligand by following the steps below.
Tools -> Surface/binding Analysis -> ViewDock -> Select the Covalent Dock output file. (covalent_out_scored.mol2) In the loaded dialog box select Dock4,5 or 6 Column -> Show -> Grid_Score Column -> Show -> HA_RMSDs
1.9 RMSD is a sampling success. Is it also a scoring success? To understand it we nee to see which of the molecules was ranked first. TO do this let's open covalent_out_scored.mol2 in text editor and see which ligand is appeared first. If it is a molecule with a lowest RMSD than it is a success, but in our case it is not. It means it is a scoring failure. The lowest RMSD structure is ranked second.