Difference between revisions of "2021 DOCK tutorial 3 with PDBID 1S19"
Stonybrook (talk | contribs) (→Footprint Analysis) |
(→Footprint Analysis) |
||
| (37 intermediate revisions by 2 users not shown) | |||
| Line 83: | Line 83: | ||
We are now left with just the ligand as pictured below. | We are now left with just the ligand as pictured below. | ||
| − | [[File:1s19_lig_woH.png|thumb|center| | + | [[File:1s19_lig_woH.png|thumb|center|400px| Figure 2. Ligand MC9 without Hydrogrens]] |
We will save this as a mol2 file by: | We will save this as a mol2 file by: | ||
| Line 95: | Line 95: | ||
After doing so, the structure should look like the one below. | After doing so, the structure should look like the one below. | ||
| − | [[File:1s19_lig_dock.png|thumb|center| | + | [[File:1s19_lig_dock.png|thumb|center|400px| Figure 3. Ligand MC9 with Hydrogrens]] |
Now we have to add the charges to the ligand. '''Remember to double check the crystal structure and validate the charge'''. In this case the charge is neutral (+/- 0), and Chimera was able to model it correctly. However this is not always the case and you will sometimes be required to manually change the charges. | Now we have to add the charges to the ligand. '''Remember to double check the crystal structure and validate the charge'''. In this case the charge is neutral (+/- 0), and Chimera was able to model it correctly. However this is not always the case and you will sometimes be required to manually change the charges. | ||
| Line 110: | Line 110: | ||
Move this to the "002.surface_spheres" directoy in the Bash Shell environment. THe following Figure is the depiction of the surface in Chimera. | Move this to the "002.surface_spheres" directoy in the Bash Shell environment. THe following Figure is the depiction of the surface in Chimera. | ||
| − | [[File:1s19_surface.png|thumb|center| | + | [[File:1s19_surface.png|thumb|center|600px| Figure 4. Receptor surface]] |
===Generating Spheres=== | ===Generating Spheres=== | ||
| Line 134: | Line 134: | ||
When this is run, the program will generate the "1s19_receptor_woH.sph" file and also an "OUTSPH" file. Looking at 1s19_receptor_woH.sph in Chimera will give you the following image. | When this is run, the program will generate the "1s19_receptor_woH.sph" file and also an "OUTSPH" file. Looking at 1s19_receptor_woH.sph in Chimera will give you the following image. | ||
| − | [[File:1s19_spheres.png|thumb|center| | + | [[File:1s19_spheres.png|thumb|center|500px| Figure 5. Generated spheres by sphgen program]] |
'''Selecting Spheres''' | '''Selecting Spheres''' | ||
| Line 142: | Line 142: | ||
This generated a new file called "selected_spheres.sph" in our working directory. The result of the sphere selection is show in the image below. | This generated a new file called "selected_spheres.sph" in our working directory. The result of the sphere selection is show in the image below. | ||
| − | [[File:1s19_selected_pheres.png|thumb|center| | + | [[File:1s19_selected_pheres.png|thumb|center|500px| Figure 6. Selected spheres within 10 angstrom of ligand and receptor]] |
= Creating Box and Grid = | = Creating Box and Grid = | ||
| Line 269: | Line 269: | ||
You can visualize the minimized ligand and the original ligand in Chimera to observe the difference between the two. | You can visualize the minimized ligand and the original ligand in Chimera to observe the difference between the two. | ||
| − | [[File:minimized.png|thumb|center| | + | [[File:minimized.png|thumb|center|400px| Figure 7. Difference in ligands after minimization]] |
The beige structure is the minimized ligand and the blue structure is the original ligand. The difference between the two is not large. To see the RMSD (room mean squared deviation) between the two, you can vim into the '''1s19.lig.min_scored.mol2''' file that was generated. The picture below shows the top of that file, and RMSD between the two molecules is 0.2281. | The beige structure is the minimized ligand and the blue structure is the original ligand. The difference between the two is not large. To see the RMSD (room mean squared deviation) between the two, you can vim into the '''1s19.lig.min_scored.mol2''' file that was generated. The picture below shows the top of that file, and RMSD between the two molecules is 0.2281. | ||
| − | [[File: | + | |
| + | [[File:1s19_minimized.png|thumb|center|600px| Figure 8. RMSD between minimized and starting ligand]] | ||
= Footprint Analysis = | = Footprint Analysis = | ||
| Line 337: | Line 338: | ||
dock6 -i footprint.in | dock6 -i footprint.in | ||
| − | To visualize the footprint, we can run a python script that will plot the results for us. | + | To visualize the footprint, we can run [http://ringo.ams.sunysb.edu/~rizzo/StonyBrook/downloads/plot_footprint_single_magnitude.py a python script] that will plot the results for us. |
| − | |||
| − | |||
With this script in our directory, we can run it by the following line. The 50 at the end of the line denotes that we are going to be plotting the distribution of the 50 most significant residues. | With this script in our directory, we can run it by the following line. The 50 at the end of the line denotes that we are going to be plotting the distribution of the 50 most significant residues. | ||
| Line 347: | Line 346: | ||
The output plot from the python script should look like the picture below. | The output plot from the python script should look like the picture below. | ||
| − | [[File:1s19_footprint.png|thumb|center| | + | [[File:1s19_footprint.png|thumb|center|400px| Figure 9. Footprint analysis from the minimiziation]] |
This process was done for the minimized ligand. However, after following the next three docking steps in the tutorial, you can repeat the footprint analysis to see the effects that changing the positioning of the ligand has on the protein. | This process was done for the minimized ligand. However, after following the next three docking steps in the tutorial, you can repeat the footprint analysis to see the effects that changing the positioning of the ligand has on the protein. | ||
| Line 429: | Line 428: | ||
This will output the file rigid.out_scored.mol2, which is visualized in Chimera (blue) over the original minimized ligand (purple). | This will output the file rigid.out_scored.mol2, which is visualized in Chimera (blue) over the original minimized ligand (purple). | ||
| − | [[File:-home-user-Documents-AMS 536 AR-1S19-004.dock-1S19 rigid-1S19 ligand rigid.jpg|thumb|center|800px|]] | + | [[File:-home-user-Documents-AMS 536 AR-1S19-004.dock-1S19 rigid-1S19 ligand rigid.jpg|thumb|center|800px| Figure 10. Rigid docked ligand (blue) compared with minimized ligand (purple)]] |
===Fixed Anchor Docking=== | ===Fixed Anchor Docking=== | ||
| Line 513: | Line 512: | ||
Upon finishing the 1S19_fixed directory should contain two new output files, fixed.out and 1S19_fad_scored.mol2, which you can open in Chimera as shown. | Upon finishing the 1S19_fixed directory should contain two new output files, fixed.out and 1S19_fad_scored.mol2, which you can open in Chimera as shown. | ||
| − | [[File:-home-user-Documents-AMS 536 AR-1S19-004.dock-1S19 fixed-1S19 docking fixed.jpg|thumb|center|800px|The original ligand position is in green with the results from the fixed docking in purple.]] | + | [[File:-home-user-Documents-AMS 536 AR-1S19-004.dock-1S19 fixed-1S19 docking fixed.jpg|thumb|center|800px| FIgure 11. The original ligand position is in green with the results from the fixed docking in purple.]] |
We can also use Chimera to see the calculated properties of the different poses for the ligand. To do this: | We can also use Chimera to see the calculated properties of the different poses for the ligand. To do this: | ||
| Line 528: | Line 527: | ||
Column -> Show -> HA_RMSDs | Column -> Show -> HA_RMSDs | ||
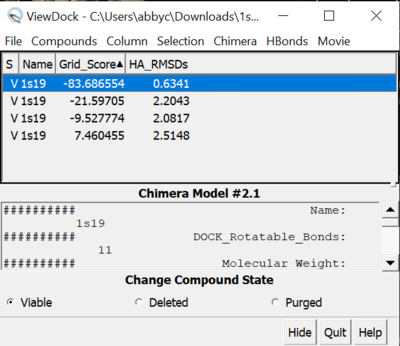
| − | [[File:1s19_viewdock.png|thumb|center| | + | [[File:1s19_viewdock.png|thumb|center|400px| Figure 12. Grid score and RMSD results from ViewDock for Fixed Anchor Docking]] |
===Flexible Docking=== | ===Flexible Docking=== | ||
| Line 614: | Line 613: | ||
rank_ligands no | rank_ligands no | ||
| − | This will output the file | + | This will output the file flex.out_scored.mol2, which is visualized in Chimera (blue) over the original minimized ligand (purple). |
| − | [[File:1S19_flex.jpg|thumb|center|800px|]] | + | [[File:1S19_flex.jpg|thumb|center|800px| Figure 13. The flex docked ligand (blue) compared to the minimized ligand (purple).]] |
= Virtual Screening = | = Virtual Screening = | ||
| Line 705: | Line 704: | ||
#!/bin/bash | #!/bin/bash | ||
#SBATCH --time=48:00:00 | #SBATCH --time=48:00:00 | ||
| − | #SBATCH --nodes= | + | #SBATCH --nodes=6 |
| − | #SBATCH --ntasks= | + | #SBATCH --ntasks=40 |
#SBATCH --job-name=1S19_VS_AR | #SBATCH --job-name=1S19_VS_AR | ||
#SBATCH --output=1S19_VS.out | #SBATCH --output=1S19_VS.out | ||
| − | #SBATCH -p long- | + | #SBATCH -p long-40core |
cd $SLURM_SUBMIT_DIR | cd $SLURM_SUBMIT_DIR | ||
echo "starting Dock6.9 simulation" | echo "starting Dock6.9 simulation" | ||
| − | /gpfs/projects/AMS536/zzz.programs/dock6.9_release/bin/dock6 -i virtual.in -o virtual.out | + | mpirun -np 240 /gpfs/projects/AMS536/zzz.programs/dock6.9_release/bin/dock6 -i virtual.in -o virtual.out |
Here we are directing SLURM to send our command to one node with one task for 48 hours on the long-28core node. You can submit the SLURM script to Seawulf using the following command: | Here we are directing SLURM to send our command to one node with one task for 48 hours on the long-28core node. You can submit the SLURM script to Seawulf using the following command: | ||
| Line 739: | Line 738: | ||
#!/bin/bash | #!/bin/bash | ||
#SBATCH --time=48:00:00 | #SBATCH --time=48:00:00 | ||
| − | #SBATCH --nodes= | + | #SBATCH --nodes=6 |
| − | #SBATCH --ntasks= | + | #SBATCH --ntasks=40 |
#SBATCH --job-name=mpi_1S19_VS_AR | #SBATCH --job-name=mpi_1S19_VS_AR | ||
#SBATCH --output=1S19_VS_mpi.out | #SBATCH --output=1S19_VS_mpi.out | ||
| Line 747: | Line 746: | ||
cd $SLURM_SUBMIT_DIR | cd $SLURM_SUBMIT_DIR | ||
echo "starting Dock6.9 simulation" | echo "starting Dock6.9 simulation" | ||
| − | /gpfs/projects/AMS536/zzz.programs/dock6.9_release/bin/dock6.mpi -i virtual.in -o virtual.out | + | mpirun -np 240 /gpfs/projects/AMS536/zzz.programs/dock6.9_release/bin/dock6.mpi -i virtual.in -o virtual.out |
Submit this to SLURM as before and wait for your job to complete. Once done, you can count how many molecules were processed using the following command: | Submit this to SLURM as before and wait for your job to complete. Once done, you can count how many molecules were processed using the following command: | ||
grep -wc "Molecule:" virtual.out | grep -wc "Molecule:" virtual.out | ||
| + | |||
| + | Now we are running the same job split into 240 separate jobs, so we should see 240 output files. Luckily all of the 5707 results will be summarized in the virtual.out file. You can open the .mol2 file in Chimera to visualize and rank your results. | ||
=Cartesian Minimization= | =Cartesian Minimization= | ||
| − | You should open the virtual.out_scored.mol2 in Chimera to visualize all of the molecules that were screened. To view and rank the molecules by properties, go to Column --> Show. | + | You should open the virtual.out_scored.mol2 in Chimera to visualize all of the molecules that were screened. To view and rank the molecules by properties, go to Tools --> Surface/Binding Analysis --> ViewDock --> Column --> Show. In the previous docking we did not minimize the ligands. We need to do this before we can make a final judgement on the best binders. |
| + | |||
| + | Input the following file into Dock6. Remember to change your directories as appropriate: | ||
| + | |||
| + | conformer_search_type rigid | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100 | ||
| + | ligand_atom_file ../007.virtual_screen_mpi/virtual.out_scored.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary yes | ||
| + | grid_score_secondary no | ||
| + | grid_score_rep_rad_scale 1 | ||
| + | grid_score_vdw_scale 1 | ||
| + | grid_score_es_scale 1 | ||
| + | grid_score_grid_prefix ../003.gridbox/grid | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_secondary no | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_secondary no | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand yes | ||
| + | simplex_max_iterations 1000 | ||
| + | simplex_tors_premin_iterations 0 | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl | ||
| + | ligand_outfile_prefix 1S19_VS_min | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| + | |||
| + | Submit this script to SLURM using the following script. Remember to change your nodes, tasks, and partition depending on your parameters: | ||
| + | |||
| + | vi job.sh | ||
| + | |||
| + | #!/bin/bash | ||
| + | #SBATCH --time=48:00:00 | ||
| + | #SBATCH --nodes=2 | ||
| + | #SBATCH --ntasks=28 | ||
| + | #SBATCH --job-name=mpi_1S19_VS_AR | ||
| + | #SBATCH --output=1S19_VS_mpi.out | ||
| + | #SBATCH -p long-28core | ||
| + | |||
| + | cd $SLURM_SUBMIT_DIR | ||
| + | echo "starting Dock6.9 simulation" | ||
| + | mpirun -np 56 /gpfs/projects/AMS536/zzz.programs/dock6.9_release/bin/dock6.mpi -i min_scored.in -o min_scored.out | ||
| + | |||
| + | = Rescoring Molecules = | ||
| + | |||
| + | Next we can rescore and rank our ligands based on footprint, Tanimoto score (synthetic accessibility, Hungarian, and volume overlap using the minimized ligand library output. First change directory: | ||
| + | |||
| + | cd 009.rescore | ||
| + | |||
| + | Then create a file called rescore.in with the following input parameters (remember to change your directory as appropriate): | ||
| + | |||
| + | conformer_search_type rigid | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file ../008.cartesianmin/1S19_VS_min_scored.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary no | ||
| + | grid_score_secondary no | ||
| + | multigrid_score_primary no | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_primary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_primary no | ||
| + | continuous_score_secondary no | ||
| + | footprint_similarity_score_primary no | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_primary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_primary yes | ||
| + | descriptor_score_secondary no | ||
| + | descriptor_use_grid_score no | ||
| + | descriptor_use_multigrid_score no | ||
| + | descriptor_use_continuous_score yes | ||
| + | descriptor_use_footprint_similarity yes | ||
| + | descriptor_use_pharmacophore_score yes | ||
| + | descriptor_use_tanimoto yes | ||
| + | descriptor_use_hungarian yes | ||
| + | descriptor_use_volume_overlap yes | ||
| + | descriptor_cont_score_rec_filename ../001.structure/1S19_rec_dockprep.mol2 | ||
| + | descriptor_cont_score_att_exp 6 | ||
| + | descriptor_cont_score_rep_exp 12 | ||
| + | descriptor_cont_score_rep_rad_scale 1 | ||
| + | descriptor_cont_score_use_dist_dep_dielectric yes | ||
| + | descriptor_cont_score_dielectric 4.0 | ||
| + | descriptor_cont_score_vdw_scale 1 | ||
| + | descriptor_cont_score_es_scale 1 | ||
| + | descriptor_fps_score_use_footprint_reference_mol2 yes | ||
| + | descriptor_fps_score_footprint_reference_mol2_filename ../004.dock/1S19_lig_min_scored.mol2 | ||
| + | descriptor_fps_score_foot_compare_type Euclidean | ||
| + | descriptor_fps_score_normalize_foot no | ||
| + | descriptor_fps_score_foot_comp_all_residue yes | ||
| + | descriptor_fps_score_receptor_filename ../001.structure/1S19_rec_dockprep.mol2 | ||
| + | descriptor_fps_score_vdw_att_exp 6 | ||
| + | descriptor_fps_score_vdw_rep_exp 12 | ||
| + | descriptor_fps_score_vdw_rep_rad_scale 1 | ||
| + | descriptor_fps_score_use_distance_dependent_dielectric yes | ||
| + | descriptor_fps_score_dielectric 4.0 | ||
| + | descriptor_fps_score_vdw_fp_scale 1 | ||
| + | descriptor_fps_score_es_fp_scale 1 | ||
| + | descriptor_fps_score_hb_fp_scale 0 | ||
| + | descriptor_fms_score_use_ref_mol2 yes | ||
| + | descriptor_fms_score_ref_mol2_filename ../004.dock/1S19_lig_min_scored.mol2 | ||
| + | descriptor_fms_score_write_reference_pharmacophore_mol2 no | ||
| + | descriptor_fms_score_write_reference_pharmacophore_txt no | ||
| + | descriptor_fms_score_write_candidate_pharmacophore no | ||
| + | descriptor_fms_score_write_matched_pharmacophore no | ||
| + | descriptor_fms_score_compare_type overlap | ||
| + | descriptor_fms_score_full_match yes | ||
| + | descriptor_fms_score_match_rate_weight 5.0 | ||
| + | descriptor_fms_score_match_dist_cutoff 1.0 | ||
| + | descriptor_fms_score_match_proj_cutoff 0.7071 | ||
| + | descriptor_fms_score_max_score 20 | ||
| + | descriptor_fingerprint_ref_filename ../004.dock/1S19_lig_min_scored.mol2 | ||
| + | descriptor_hms_score_ref_filename ../004.dock/1S19_lig_min_scored.mol2 | ||
| + | descriptor_hms_score_matching_coeff -5 | ||
| + | descriptor_hms_score_rmsd_coeff 1 | ||
| + | descriptor_volume_score_reference_mol2_filename ../004.dock/1S19_lig_min_scored.mol2 | ||
| + | descriptor_volume_score_overlap_compute_method analytical | ||
| + | descriptor_weight_cont_score 1 | ||
| + | descriptor_weight_fps_score 1 | ||
| + | descriptor_weight_pharmacophore_score 1 | ||
| + | descriptor_weight_fingerprint_tanimoto -1 | ||
| + | descriptor_weight_hms_score 1 | ||
| + | descriptor_weight_volume_overlap_score -1 | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl | ||
| + | chem_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/chem.defn | ||
| + | pharmacophore_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/ph4.defn | ||
| + | ligand_outfile_prefix descriptor.output | ||
| + | write_footprints yes | ||
| + | write_hbonds yes | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| + | |||
| + | Use the same ViewDock tool in Chimera to see the different descriptors for each ligand. | ||
| + | |||
| + | = Visualizing Virtual Screen Results = | ||
| + | Now that we've successfully run our virtual screen, we want to view our results. We can do this using Chimera. In Chimera, we are going to load in the following files: '''1s19.box.pdb, selected_spheres.sph, 1s19_dockprep.mol2, 1s19_ligand_dockprep.mol2 and virtual.out_scored.mol2'''. It is important to note that we are going to be using '''ViewDock''' to open our virtual screen. To do this: | ||
| + | |||
| + | Tools -> Structure/Binding Analysis -> ViewDock | ||
| + | Follow the prompts to open your virtual.out_scored.mol2 | ||
| + | Make sure to select Dock 4,5,6 when asked | ||
| + | |||
| + | Once opened in Chimera, you will see an image like the one below: | ||
| + | |||
| + | [[File:1s19_virtualscreen.png|thumb|center|500px| Figure 14. The loaded virtual screen results in Chimera.]] | ||
| + | |||
| + | Now we can do some analysis with this Chimera session. To start, we are going to: | ||
| + | |||
| + | Favorites -> Model Panel | ||
| + | |||
| + | In '''Model Panel''' we can control the visibility of each of the files that we have loaded in. For now, we are going to uncheck our box, selected spheres and the protein in the '''Shown''' category. This will hide those three files to make it easier to visualize. To rank our virtual screen results, we are going to use our ViewDock window. In ViewDock: | ||
| + | |||
| + | Column -> Show -> Grid_Score | ||
| + | |||
| + | Once this panel opens next to the ligands, you can click on the title of the column to organize the ligands from lowest grid score to highest. Selecting one of the ligands will display one of the ligands at a time, and you can push the down arrow on your keyboard to scroll through all of the virtual screen results to visualize them. | ||
| + | |||
| + | We can also visualize the hydrogen bonds that each of the ligands creates with the surrounding residues of the protein. To do this: | ||
| + | |||
| + | Tools -> Structure/Binding Analysis -> FindHBond | ||
| + | |||
| + | In the HBond window we are going to make a few adjustments. First, in the upper right hand box, we are going to make sure that '''inter-model''' is checked off. We do not want to find hbonds between residues of the protein or ligands, but between each other. Next, you can choose the HBond color at the top (we use magenta so it is easy to see) and set the line thickness to 4.0. We are going to keep the '''Relax H-Bond constraints''' box checked. | ||
| + | |||
| + | Next, we are going to check off the '''Restrict to Models''' box. When we do this, we will see a list of all of the models that we currently have loaded into Chimera. For our first HBond analysis, we are going to select all of our virtual screen ligands and the protein. To do this, select the first ligand at the top and hold the '''shift''' key to select the rest. Once they are all selected, we are going to go ahead and check off '''If endpoint atom hidden, show endpoint residue'''. This will show only the residues of the protein that are making HBonds with the ligand - this makes the visualization in Chimera much less crammed. | ||
| + | |||
| + | Now we can go back to '''Model Panel''' and scroll through all of our virtual screen ligands again to view the different HBonds that they make within the binding pocket of the protein. To compare the HBonds that the ligands make to our original ligand, we are going to repeat the HBond analysis process for the ligand. We are going to keep all of the parameters in the HBond window the same as before, except now we are only going to select our dockprepped ligand. We are also going to check off the box '''Retain currently displayed H-Bonds''' so we do not lose the HBond data from all of the virtual screen ligands. | ||
| + | |||
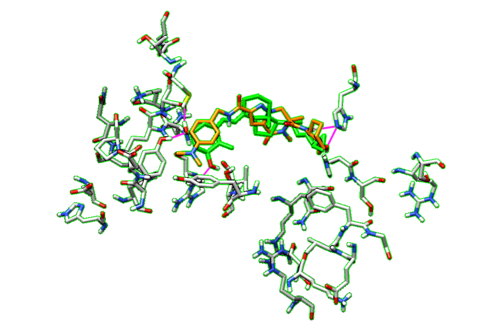
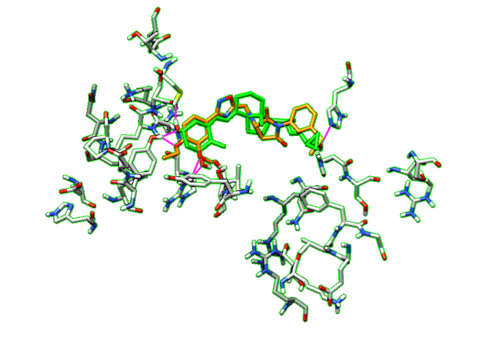
| + | With the dockprepped ligand's HBonds shown now, now we can scroll back through our list of ligands and see how their HBonds compare to our original ligand. Some examples of what this should look like are in the figure below. | ||
| + | |||
| + | [[File:1s19_hbonds1.png|thumb|center|500px| Figure 15. Example of Hbonds in Chimera.]] | ||
| + | [[File:1s19_hbonds2.png|thumb|center|500px| Figure 16. Example of Hbonds in Chimera.]] | ||
Latest revision as of 12:42, 7 March 2022
Contents
Introduction
DOCK
Developed by Irwin D. Kuntz, Jr. and colleagues at UCSF, DOCK is a program used to dock molecules to one another. Docking is a process in which given a small molecule or ligand in the active site of a receptor, the program will try to predict the lowest-energy binding mode of the ligand to the receptor. This process is very important in drug discovery as small molecules that bind to or interact with the active site of a receptor associated with a disease could inhibit its function, acting as a drug. DOCK is a particularly helpful tool as it is used to screen massive libraries of molecules, containing millions of compounds, against a receptor to identify the most promising drug lead compounds.
DOCK historically used a geometric matching algorithm to superimpose the ligand onto a negative image of the active site of the receptor. Over the years, features were added that would improve the programs ability to find the lowest-energy binding mode. Somem of these features include force-field based scoring, on-the-fly optimization and an algorithm for flexible ligand docking.
In this tutorial DOCK6 will be used. New features for DOCK6 include: additional scoring options during minimization; DOCK 3.5 scoring-including Delphi electrostatics, ligand conformational entropy corrections, ligand desolvation, receptor desolvation; Hawkins-Cramer-Truhlar GB/SA solvation scoring with optional salt screening and more (see UCSF DOCK). These new features improved the programs ability to predict binding poses.
1S19
This tutorial will use PDB code: 1S19. 1S19 is the crystal structure of VDR ligand binding domain complexed to calcipotriol (find structure here. The resolution is 2.10 Å.
Chimera
UCSF Chimera was developed by Resource for Biocomputing, Visualization, and Informatics (RBVI) at the University of California, San Francisco. Chimera is a program made for the interactive visualization and analysis of molecular structures. Some features of Chimera include general structure analysis (automatic identification of an atom, hydrogen addition and partial charge assignment, structure building and bond rotation, etc.) presenting images and movies (high-res images, visual effects, standard molecular representations, etc.) and sequence structure tools (sequence alignments, structure superposition, etc.)
Making Directories
To make it easier for us to locate certain files, we are going to create directories for each step of the docking process. The mkdir command creates a new directory in which new files can be saved. The cd command allows for you to navigate between directories.
In your Bash Shell environment, create a directory containing all of the information for the project by typing:
mkdir 1S19
To change into that directory type:
cd 1S19
To create the directories for each step:
mkdir 001.structure 002.surface_spheres 003.gridbox 004.dock 005.virtual_screen 006.virtual_screen_mpi 007.cartesianmin 008.rescore
To confirm that the directories have been created:
ls
If you made a mistake and need to delete a directory:
rm *insert name of directory to be deleted*
Preparing the Ligand and Receptor
PDB Structure
Download the structure of 1S19 from the Protein Data Bank (PDB).
Download Files -> PDB Format
This file contains the coordinates for the 3D structure of the receptor, ligand and any other molecules present during the experiment (typically water or metal ions). To visualize the structure, we will be using Chimera.
Visualization with Chimera To open the newly downloaded PDB coordinates:
File -> Open
The protein when you first open the file should look like the image below. You can change the view of the structure by rotating it with your mouse or touchpad. Currently, some of the side chains of the backbone are shown, there are no hydrogens or partial charges, and there are some water molecules present.
Receptor
In order to dock, the ligand and receptor have to be separated and saved into different files. To do this is a simple process and can be done in Chimera. To hide all sidechain bonds:
Select -> standard amino acids Actions -> Atoms/Bonds -> hide
To prepare the receptor, we are going to want to delete everything except the protein from the PDB file. To do this in Chimera:
Select -> Residue -> All nonstandard Actions -> Atoms/Bonds -> Delete
You will now be left with only the desired protein. To save this file, we are going to save it as a mol2 file. This can be done by:
File -> Save Mol2 -> "1s19_receptor_woH.mol2"
Adding Hydrogens and Charge PDB structures are reported without hydrogens, so it is important that we add them to the receptor in order to gain accurate calculations for the interactions between the protein and ligand. This can also be done in Chimera by doing the following:
Tools -> Structure Editing -> Add H -> Ok
We will also need to add charges to the receptor
Tools -> Structure Editing -> Add Charge -> (have Amber ff14SB and AM1-BCC selected) -> Ok
Once this is done it is very important to check that the charges of the receptor match that of the experiments. Chimera adds the standard protonation states to the amino acids, so it's important to read the paper associated with the PDB file (see here) to make sure that there are no amino acids that are specifically protonated or deprotonated.
Once you have checked to make sure that the protonation states are okay, save this as a mol2 file:
File -> Save Mol2 -> "1s19_receptor_dockprep.mol2"
Ligand
Like the receptor, we will need to save the ligand as a separate mol2 file in order to perform the docking. For this model, the ligand is named as MC9 in Chimera.
To isolate the ligand:
Select -> Residue -> MC9 Select -> Invert (all models) Actions -> Atoms/Bonds -> Delete
We are now left with just the ligand as pictured below.
We will save this as a mol2 file by:
File -> Save Mol2 -> 1s19_lig_woH.mol2
Adding Hydrogens and Charges In the same steps as the receptor, we will add hydrogens and partial charges to the ligand. For the Hydrogens:
Tools -> Structure Editing -> Add H
After doing so, the structure should look like the one below.
Now we have to add the charges to the ligand. Remember to double check the crystal structure and validate the charge. In this case the charge is neutral (+/- 0), and Chimera was able to model it correctly. However this is not always the case and you will sometimes be required to manually change the charges.
Save this final structure as a mol2 file with the name "1s19_lig_dockprep.mol2"
Generating Receptor Surface and Spheres
Generating Surface
In Chimera we can generate a representation of the receptor that creates a negative image of the protein. This surface will guide the ligand to the active site of the receptor during docking. To do this, we are going to open the receptor without hydrogens in Chimera, 1s19_receptor_woH.mol2. Then in Chimera, do:
Actions -> Surface -> Show Tools -> Structure Editing -> Write DMS -> "1s19_rec_surface.dms"
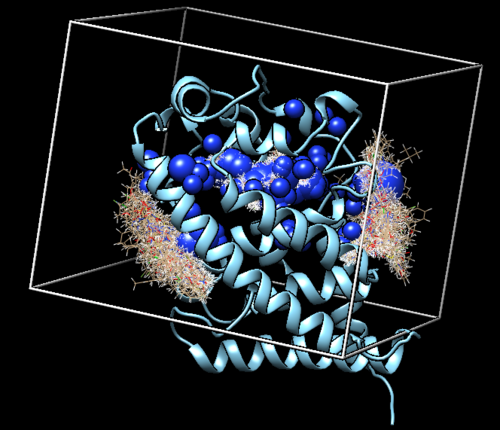
Move this to the "002.surface_spheres" directoy in the Bash Shell environment. THe following Figure is the depiction of the surface in Chimera.
Generating Spheres
Spheres in docking represent empty space in the receptor. Now that we have the surface representation of our receptor, we are going to use the sphgen ("sphere generation") script to create the largest possible sphere for any given empty space. It is okay that spheres overlap with each other, however we do not want any spheres overlapping with the protein.
Still in the "002.surface_spheres" directory, we are going to create a file calle INSPH (short for "in spheres"). To do this type:
vim INSPH
In this new file, we are going to write a script in order to automate the process of generating the spheres. Below is an example of the script that we used.
1s19_rec_surface.dms # your receptor as DMS file R # <R flag> - enables sphere generation outside the protein surface (no eclipsing) X # <X flag - uses all coordinates 0.0 # <double> - distance that steric interactions are checked 4.0 # <double> - Maximum sphere radius of generated sphere 1.4 # <double> - Size of sphere that rolls over dms file surface for cavities 1s19_receptor_woH.sph # the name of your output file
With this script saved, you can now run it through sphgen by the following:
sphgen -i INSPH -o OUTSPH
When this is run, the program will generate the "1s19_receptor_woH.sph" file and also an "OUTSPH" file. Looking at 1s19_receptor_woH.sph in Chimera will give you the following image.
Selecting Spheres We are more concerned with the empty space near the active site of the receptor, since that is where we are going to be docking our ligand. To only select particular spheres within 10 angstroms of the ligand, we executed the following command:
sphere_selector 3vjk_receptor_woH.sph 3vjk_ligand_H.mol2 10.0
This generated a new file called "selected_spheres.sph" in our working directory. The result of the sphere selection is show in the image below.
Creating Box and Grid
To make the amount of calculations we have to do smaller, and thus more efficient, we will create a grid in which relevent residues/atoms are selected. This method of creating the box and grid will eliminate any long-distance interactions that the ligand may have.
Generating Box
We will create the box by using the Showbox program. To do this, we will create the following file in our "003.gridbox" directory.
vim showbox.in
We put the following script inside the showbox.in file:
Y #generate box# 8.0 #how many angstroms the box edges should be from the spheres ./../002.surface_spheres/select_spheres.sph #the location of the selected spheres 1 1s19.box.pdb #name of the output file
To execute this script, type the following:
showbox < showbox.in
Once this program has run, the 1s19.box.pdb file should be in the working directory.
Generating Grid
Next, we will be generating the grid. To do so, we will create a file called grid.in
vim grid.in
The following script will be copied into the grid.in file
compute_grids yes grid_spacing 0.4 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4. bump_filter yes bump_overlap 0.75 receptor_file ./../001.structure/1s19_receptor_dockprep.mol2 box_file 1s19.box.pbd vdw_definition_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
To run this script, type:
grid -i grid.in -o gridinfo.out
When this program finishes running, you should have three new files: gridinfo.out, grid.nrg, and grid.bmp
Energy Minimization
The ligand pose in the crystal structure may not accurately represent the lowest energy conformation of the ligand in a biological system. Therefore, to accurately dock the ligand, we first need to reduce the overall potential energy of the ligand to its global minimum. To do this, we are going to create an energy minimization script:
vim min.in
We used the following script for the minimizaiton:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ./../001.build/1s19_ligand_dockprep.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ./../001.build/1s19_ligand_dockprep.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ./../003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix 1s19.lig.min write_orientations no num_scored_conformers 1 rank_ligands no
Now that the script is made, we can run the energy minimization by the following line
dock6 -i min.in -o min.out
Running this command will yield two new files in the direectory: min.out and 1s19.lig.min_scored.mol2
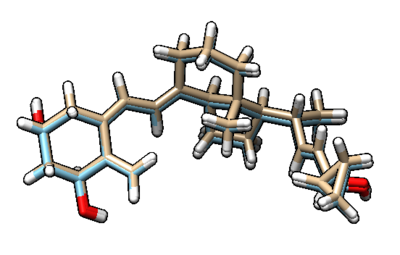
You can visualize the minimized ligand and the original ligand in Chimera to observe the difference between the two.
The beige structure is the minimized ligand and the blue structure is the original ligand. The difference between the two is not large. To see the RMSD (room mean squared deviation) between the two, you can vim into the 1s19.lig.min_scored.mol2 file that was generated. The picture below shows the top of that file, and RMSD between the two molecules is 0.2281.
Footprint Analysis
We can use the moleucular footprint to understand the lignad binding to the receptor by using electrostatic and Van der Waals interactions. We will be doing footprint analysis for our minimization of the ligand and also the rigid, flex, and fixed anchor docking. To begin this analysis, create the file footprint.in and copy the following script into the file:
conformer_search_type rigid use_internal_energy no ligand_atom_file ./../004.dock/1s19.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary yes footprint_similarity_score_secondary no fps_score_use_footprint_reference_mol2 yes fps_score_footprint_reference_mol2_filename ./../001.structure/1s19_ligand_dockprep.mol2 fps_score_foot_compare_type Euclidean fps_score_normalize_foot no fps_score_foot_comp_all_residue yes fps_score_receptor_filename ./../001.structure/1s19_dockprep.mol2 fps_score_vdw_att_exp 6 fps_score_vdw_rep_exp 9 fps_score_vdw_rep_rad_scale 1 fps_score_use_distance_dependent_dielectric yes fps_score_dielectric 4.0 fps_score_vdw_fp_scale 1 fps_score_es_fp_scale 1 fps_score_hb_fp_scale 0 pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix footprint.out write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
To run the script, type the following:
dock6 -i footprint.in
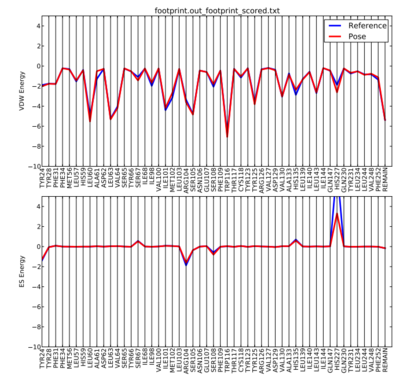
To visualize the footprint, we can run a python script that will plot the results for us.
With this script in our directory, we can run it by the following line. The 50 at the end of the line denotes that we are going to be plotting the distribution of the 50 most significant residues.
python plot_footprint_single_magnitude.py footprint.out_footprint_scored.txt 50
The output plot from the python script should look like the picture below.
This process was done for the minimized ligand. However, after following the next three docking steps in the tutorial, you can repeat the footprint analysis to see the effects that changing the positioning of the ligand has on the protein.
Docking
Rigid Docking
Rigid is the least computationally intensive form of docking - the ligand has no rotational freedom around its bonds and is kept static. The first step is to move to the dock folder and make a folder for rigid docking.
cd 004.dock mkdir 1S19_rigid
Then we can make an input file for rigid docking and edit in within Vim or the Dock6.9 program.
touch rigid.in vim rigid.in
Here we will edit the parameters as follows:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100 ligand_atom_file ./../004.dock/1S19_lig_min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ./../004.dock/1S19_lig_min_scored.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ./../002.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ./../003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix rigid.out write_orientations no num_scored_conformers 1 rank_ligands no
Then we will input the file into Dock6.9 with the following command:
dock6 -i rigid.in
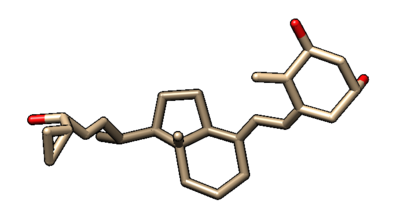
This will output the file rigid.out_scored.mol2, which is visualized in Chimera (blue) over the original minimized ligand (purple).
Fixed Anchor Docking
Fixed docking is the next level up. Here we will keep the major part of the ligand with no rotatable bonds fixed as an anchor but allow rotational freedom around the other rigid groups. Let's begin by making a directory and input file within 004.dock:
cd 004.dock mkdir 1S19_fixed cd 1S19_fixed touch fixed.in
This file can be modified in Vim or in Dock6.9 itself as shown:
conformer_search_type flex write_fragment_libraries no user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100 pruning_conformer_score_scaling_factor 1 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100 ligand_atom_file ../../001.structure/1S19_ligand_dockprep.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../../001.structure/1S19_ligand_dockprep.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../../003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1 simplex_trans_step 1 simplex_rot_step 0.1 simplex_tors_step 10 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix 1S19_fad write_orientations no num_scored_conformers 100 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
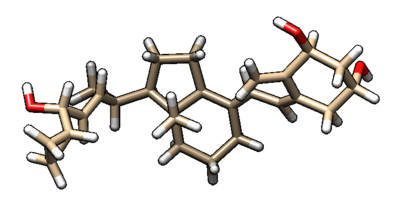
Upon finishing the 1S19_fixed directory should contain two new output files, fixed.out and 1S19_fad_scored.mol2, which you can open in Chimera as shown.
We can also use Chimera to see the calculated properties of the different poses for the ligand. To do this:
File -> Open -> 2s19_rec_dockprep.mol2 File -> Open -> 2p16_ligand_dockprep.mol2 File -> Open -> 1s19_fad_scored.mol2 Tools -> Surface/binding Analysis -> ViewDock -> 1s19_fad_scored.mol2 In the loaded dialog box select Dock4,5 or 6
Now you can select which properties of the docking you would like to see. We are going to look at grid scores and RMSDs. To do this:
Column -> Show -> Gridscore Column -> Show -> HA_RMSDs
Flexible Docking
Finally we have the most computationally intensive type of docking, flexible docking, where the ligand has full rotational freedom around its bond angles and all internal degrees of freedom are sampled. Same as before, let's make a directory for the files and write an input file:
cd 004.dock mkdir 1S19_flexible cd 1S19_flexible touch flex.in
Edit the input file in vim or in Dock6.9 itself as follows:
conformer_search_type flex write_fragment_libraries no user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100 pruning_conformer_score_scaling_factor 1 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100 ligand_atom_file ../1S19_lig_min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../1S19_lig_min_scored.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../../002.surface_spheres/selected_spheres.sph max_orientations 100 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../../003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1 simplex_trans_step 1 simplex_rot_step 0.1 simplex_tors_step 10 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix flex.out write_orientations no num_scored_conformers 1 rank_ligands no
This will output the file flex.out_scored.mol2, which is visualized in Chimera (blue) over the original minimized ligand (purple).
Virtual Screening
Virtual Screening
Virtual screening involves docking a library of ligands against a receptor and ranking them for further analysis. The library can be defined by the user based on prior knowledge of ligand size, side-groups, and favorable interactions. For this tutorial, we will be using the tutorial file "VS_library_5K.mol2" which should be copied to the 006.virtualscreen and 007.virtualscreenmpi directories.
Enter the 006.virtualscreen directory and create a Dock6 input file:
cd 006.virtualscreen touch virtual.in dock6 -i virtual.in
write_fragment_libraries no user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 9 internal_energy_cutoff 100 ligand_atom_file VS_library_5K.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix virtual.out write_orientations no num_scored_conformers 1 rank_ligands no
Running this dock process would be too computationally and time-intensive for a local computer. Therefore, we will be using SLURM to send our job to the Seawulf HPC. Write the following Bash script for SLURM submission:
touch virtual_screen.sh
#!/bin/bash #SBATCH --time=48:00:00 #SBATCH --nodes=6 #SBATCH --ntasks=40 #SBATCH --job-name=1S19_VS_AR #SBATCH --output=1S19_VS.out #SBATCH -p long-40core
cd $SLURM_SUBMIT_DIR echo "starting Dock6.9 simulation" mpirun -np 240 /gpfs/projects/AMS536/zzz.programs/dock6.9_release/bin/dock6 -i virtual.in -o virtual.out
Here we are directing SLURM to send our command to one node with one task for 48 hours on the long-28core node. You can submit the SLURM script to Seawulf using the following command:
sbatch virtual_screen.sh
You can check on the statues of your submission with the following command:
squeue -u NETID
This command will give you the JobID and runtime of your submission. To cancel the submission, use the following:
scancel JOBID
This job will take too long on one node of the HPC. It would be more efficient to run the job in parallel on multiple cores. To accomplish this, we can use MPI, AKA Message Passing Interface.
Virtual Screening with MPI
cd 007.virtualscreenmpi
Copy the VS_library_5K.mol2 and virtual.in files to the new directory. We will be using these same input files. However, we need to modify the SLURM input file to account for the new nodes.
vim virtual_screen_mpi.sh
#!/bin/bash #SBATCH --time=48:00:00 #SBATCH --nodes=6 #SBATCH --ntasks=40 #SBATCH --job-name=mpi_1S19_VS_AR #SBATCH --output=1S19_VS_mpi.out #SBATCH -p long-40core
cd $SLURM_SUBMIT_DIR echo "starting Dock6.9 simulation" mpirun -np 240 /gpfs/projects/AMS536/zzz.programs/dock6.9_release/bin/dock6.mpi -i virtual.in -o virtual.out
Submit this to SLURM as before and wait for your job to complete. Once done, you can count how many molecules were processed using the following command:
grep -wc "Molecule:" virtual.out
Now we are running the same job split into 240 separate jobs, so we should see 240 output files. Luckily all of the 5707 results will be summarized in the virtual.out file. You can open the .mol2 file in Chimera to visualize and rank your results.
Cartesian Minimization
You should open the virtual.out_scored.mol2 in Chimera to visualize all of the molecules that were screened. To view and rank the molecules by properties, go to Tools --> Surface/Binding Analysis --> ViewDock --> Column --> Show. In the previous docking we did not minimize the ligands. We need to do this before we can make a final judgement on the best binders.
Input the following file into Dock6. Remember to change your directories as appropriate:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100 ligand_atom_file ../007.virtual_screen_mpi/virtual.out_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix 1S19_VS_min write_orientations no num_scored_conformers 1 rank_ligands no
Submit this script to SLURM using the following script. Remember to change your nodes, tasks, and partition depending on your parameters:
vi job.sh
#!/bin/bash #SBATCH --time=48:00:00 #SBATCH --nodes=2 #SBATCH --ntasks=28 #SBATCH --job-name=mpi_1S19_VS_AR #SBATCH --output=1S19_VS_mpi.out #SBATCH -p long-28core
cd $SLURM_SUBMIT_DIR echo "starting Dock6.9 simulation" mpirun -np 56 /gpfs/projects/AMS536/zzz.programs/dock6.9_release/bin/dock6.mpi -i min_scored.in -o min_scored.out
Rescoring Molecules
Next we can rescore and rank our ligands based on footprint, Tanimoto score (synthetic accessibility, Hungarian, and volume overlap using the minimized ligand library output. First change directory:
cd 009.rescore
Then create a file called rescore.in with the following input parameters (remember to change your directory as appropriate):
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../008.cartesianmin/1S19_VS_min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary no footprint_similarity_score_secondary no pharmacophore_score_primary no pharmacophore_score_secondary no descriptor_score_primary yes descriptor_score_secondary no descriptor_use_grid_score no descriptor_use_multigrid_score no descriptor_use_continuous_score yes descriptor_use_footprint_similarity yes descriptor_use_pharmacophore_score yes descriptor_use_tanimoto yes descriptor_use_hungarian yes descriptor_use_volume_overlap yes descriptor_cont_score_rec_filename ../001.structure/1S19_rec_dockprep.mol2 descriptor_cont_score_att_exp 6 descriptor_cont_score_rep_exp 12 descriptor_cont_score_rep_rad_scale 1 descriptor_cont_score_use_dist_dep_dielectric yes descriptor_cont_score_dielectric 4.0 descriptor_cont_score_vdw_scale 1 descriptor_cont_score_es_scale 1 descriptor_fps_score_use_footprint_reference_mol2 yes descriptor_fps_score_footprint_reference_mol2_filename ../004.dock/1S19_lig_min_scored.mol2 descriptor_fps_score_foot_compare_type Euclidean descriptor_fps_score_normalize_foot no descriptor_fps_score_foot_comp_all_residue yes descriptor_fps_score_receptor_filename ../001.structure/1S19_rec_dockprep.mol2 descriptor_fps_score_vdw_att_exp 6 descriptor_fps_score_vdw_rep_exp 12 descriptor_fps_score_vdw_rep_rad_scale 1 descriptor_fps_score_use_distance_dependent_dielectric yes descriptor_fps_score_dielectric 4.0 descriptor_fps_score_vdw_fp_scale 1 descriptor_fps_score_es_fp_scale 1 descriptor_fps_score_hb_fp_scale 0 descriptor_fms_score_use_ref_mol2 yes descriptor_fms_score_ref_mol2_filename ../004.dock/1S19_lig_min_scored.mol2 descriptor_fms_score_write_reference_pharmacophore_mol2 no descriptor_fms_score_write_reference_pharmacophore_txt no descriptor_fms_score_write_candidate_pharmacophore no descriptor_fms_score_write_matched_pharmacophore no descriptor_fms_score_compare_type overlap descriptor_fms_score_full_match yes descriptor_fms_score_match_rate_weight 5.0 descriptor_fms_score_match_dist_cutoff 1.0 descriptor_fms_score_match_proj_cutoff 0.7071 descriptor_fms_score_max_score 20 descriptor_fingerprint_ref_filename ../004.dock/1S19_lig_min_scored.mol2 descriptor_hms_score_ref_filename ../004.dock/1S19_lig_min_scored.mol2 descriptor_hms_score_matching_coeff -5 descriptor_hms_score_rmsd_coeff 1 descriptor_volume_score_reference_mol2_filename ../004.dock/1S19_lig_min_scored.mol2 descriptor_volume_score_overlap_compute_method analytical descriptor_weight_cont_score 1 descriptor_weight_fps_score 1 descriptor_weight_pharmacophore_score 1 descriptor_weight_fingerprint_tanimoto -1 descriptor_weight_hms_score 1 descriptor_weight_volume_overlap_score -1 gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl chem_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/chem.defn pharmacophore_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/ph4.defn ligand_outfile_prefix descriptor.output write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
Use the same ViewDock tool in Chimera to see the different descriptors for each ligand.
Visualizing Virtual Screen Results
Now that we've successfully run our virtual screen, we want to view our results. We can do this using Chimera. In Chimera, we are going to load in the following files: 1s19.box.pdb, selected_spheres.sph, 1s19_dockprep.mol2, 1s19_ligand_dockprep.mol2 and virtual.out_scored.mol2. It is important to note that we are going to be using ViewDock to open our virtual screen. To do this:
Tools -> Structure/Binding Analysis -> ViewDock Follow the prompts to open your virtual.out_scored.mol2 Make sure to select Dock 4,5,6 when asked
Once opened in Chimera, you will see an image like the one below:
Now we can do some analysis with this Chimera session. To start, we are going to:
Favorites -> Model Panel
In Model Panel we can control the visibility of each of the files that we have loaded in. For now, we are going to uncheck our box, selected spheres and the protein in the Shown category. This will hide those three files to make it easier to visualize. To rank our virtual screen results, we are going to use our ViewDock window. In ViewDock:
Column -> Show -> Grid_Score
Once this panel opens next to the ligands, you can click on the title of the column to organize the ligands from lowest grid score to highest. Selecting one of the ligands will display one of the ligands at a time, and you can push the down arrow on your keyboard to scroll through all of the virtual screen results to visualize them.
We can also visualize the hydrogen bonds that each of the ligands creates with the surrounding residues of the protein. To do this:
Tools -> Structure/Binding Analysis -> FindHBond
In the HBond window we are going to make a few adjustments. First, in the upper right hand box, we are going to make sure that inter-model is checked off. We do not want to find hbonds between residues of the protein or ligands, but between each other. Next, you can choose the HBond color at the top (we use magenta so it is easy to see) and set the line thickness to 4.0. We are going to keep the Relax H-Bond constraints box checked.
Next, we are going to check off the Restrict to Models box. When we do this, we will see a list of all of the models that we currently have loaded into Chimera. For our first HBond analysis, we are going to select all of our virtual screen ligands and the protein. To do this, select the first ligand at the top and hold the shift key to select the rest. Once they are all selected, we are going to go ahead and check off If endpoint atom hidden, show endpoint residue. This will show only the residues of the protein that are making HBonds with the ligand - this makes the visualization in Chimera much less crammed.
Now we can go back to Model Panel and scroll through all of our virtual screen ligands again to view the different HBonds that they make within the binding pocket of the protein. To compare the HBonds that the ligands make to our original ligand, we are going to repeat the HBond analysis process for the ligand. We are going to keep all of the parameters in the HBond window the same as before, except now we are only going to select our dockprepped ligand. We are also going to check off the box Retain currently displayed H-Bonds so we do not lose the HBond data from all of the virtual screen ligands.
With the dockprepped ligand's HBonds shown now, now we can scroll back through our list of ligands and see how their HBonds compare to our original ligand. Some examples of what this should look like are in the figure below.