Difference between revisions of "2019 Covalent docking tutorial 1 with PDB 5VKG"
Stonybrook (talk | contribs) (→II. Ligand Preparation) |
Stonybrook (talk | contribs) (→III. Preparing the sphere files for orienting) |
||
| Line 76: | Line 76: | ||
== '''III. Preparing the sphere files for orienting''' == | == '''III. Preparing the sphere files for orienting''' == | ||
| + | We need to prepare two types of spheres: 1) spheres for box generating; 2) spheres for orienting the ligand. | ||
| − | |||
| + | |||
| + | The only spheres that will be orientated will be the alpha carbon, beta carbon, and the sulfur on the cysteine sidechain. Open the mol2 file with prepared ligand. Isolate the sulfur, alpha and beta carbons and delete everything else. | ||
[[Image:Image_n16_spheres.png|thumb|center|600px| The three atoms from the cysteine sidechain needed for oritenting: beta carbon, alpha carbon, and sulfur.]] | [[Image:Image_n16_spheres.png|thumb|center|600px| The three atoms from the cysteine sidechain needed for oritenting: beta carbon, alpha carbon, and sulfur.]] | ||
| + | |||
| + | Once that is isolated save it as a pdb or as a mol2. Create file mol2_to_sph.py and write the following into it: | ||
| + | import sys, math, mol2 | ||
| + | import os.path | ||
| + | import cmath | ||
| + | from math import sqrt | ||
| + | |||
| + | ## written by Trent E. Balius and editted by Lauren Prentis | ||
| + | ## Sept 2019 | ||
| + | ## converts a mol2 into a sph for covalent | ||
| + | |||
| + | def write_sph_file(filename,mol): | ||
| + | ##dt = mol2.convert_sybyl_to_dock(mol) # get dock atom types. | ||
| + | vdw_dict = 0.5 | ||
| + | outsph = open(filename, 'w') | ||
| + | print (len(mol.atom_list)) | ||
| + | outsph.write("DOCK spheres generated from ligand heavy atoms\n") | ||
| + | outsph.write("cluster 1 number of spheres in cluster %d\n" % len(mol.atom_list)) | ||
| + | for i in range(len(mol.atom_list)): | ||
| + | radius = 0.5 | ||
| + | outsph.write("%5d%10.5f%10.5f%10.5f%8.3f%5d 0 0\n" % | ||
| + | (i+1,round(mol.atom_list[i].X,3), | ||
| + | round(mol.atom_list[i].Y,3), | ||
| + | round(mol.atom_list[i].Z,3),radius,i+1) ) | ||
| + | outsph.close() | ||
| + | return | ||
| + | |||
| + | def remove_hydrogens(m): | ||
| + | atom_list = [] | ||
| + | bond_list = [] | ||
| + | residue_list = {} | ||
| + | |||
| + | # Retain only heavy atoms in atom_list | ||
| + | num_hvy_atoms = 0 | ||
| + | for i in range(len(m.atom_list)): | ||
| + | if (m.atom_list[i].heavy_atom): | ||
| + | atom_list.append(m.atom_list[i]) | ||
| + | num_hvy_atoms+=1 | ||
| + | |||
| + | # Retain only bonds containing heavy atoms | ||
| + | for bond_id in range(len(m.bond_list)): | ||
| + | retain_bond = True | ||
| + | for atom_id in range(len(m.atom_list)): | ||
| + | if (m.atom_list[atom_id].heavy_atom): | ||
| + | continue | ||
| + | # Atoms down here are always hydrogen | ||
| + | if (m.bond_list[bond_id].a1_num == m.atom_list[atom_id].num): | ||
| + | retain_bond = False | ||
| + | if (m.bond_list[bond_id].a2_num == m.atom_list[atom_id].num): | ||
| + | retain_bond = False | ||
| + | if (retain_bond): | ||
| + | bond_list.append(m.bond_list[bond_id]) | ||
| + | |||
| + | # Assuming that residue list does not change | ||
| + | |||
| + | data = Mol(m.header,m.name,atom_list,bond_list,m.residue_list) | ||
| + | return data | ||
| + | |||
| + | def main(): | ||
| + | if len(sys.argv) !=3: | ||
| + | print("Not enough inputs") | ||
| + | name_of_mol2 = sys.argv[1] | ||
| + | name_of_sph = sys.argv[2] | ||
| + | mol = mol2.remove_hydrogens(mol2.read_Mol2_file(name_of_mol2)[0]) | ||
| + | write_sph_file(name_of_sph,mol) | ||
| + | main() | ||
| + | |||
| + | |||
| + | |||
| + | Use a code below to convert this pdb or mol2 into a sphere file. | ||
| + | python mol2_to_sph.py lig_abs.mol2 lig_abs_spheres.sph | ||
| + | |||
Now new spheres will be generated for the box. To accomplish this spheres will be generated ligand file. A sphere file will be made using the coordinates from the ligand file of the heavy atom. Now that the spheres are generated twice once for orientating and the other for the dock/grid. The box will then be generated, once the box is made the grid can be populated in the box. Now that the grid, the ligand, and spheres meant of orientating. You can begin covalent docking. | Now new spheres will be generated for the box. To accomplish this spheres will be generated ligand file. A sphere file will be made using the coordinates from the ligand file of the heavy atom. Now that the spheres are generated twice once for orientating and the other for the dock/grid. The box will then be generated, once the box is made the grid can be populated in the box. Now that the grid, the ligand, and spheres meant of orientating. You can begin covalent docking. | ||
Revision as of 10:56, 11 March 2020
This tutorial teaches you how to dock a covalently bound drug molecule to a receptor (PDB 5VKG).
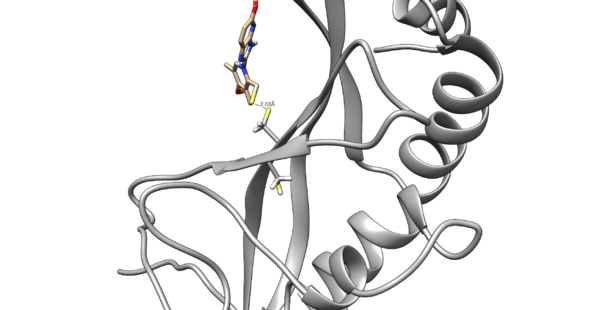
Use chimera to visualize your system in this case the pdb 5VKG of NMR structure of human Tsg101 UEV in complex with tenatoprazole. PDB 5VKG consists of 20 different models, but we need only one. Either chose one of the models or ask Lauren to give you the file rcsb104645_model1.pdb
We are interested in the interaction between S atom in Cys 73 and S atom in the ligand. Those two atoms are covalently bonded according to the experimental paper, but Chimera will not show a covalent linker between these two S. We can create the bond ourself in Chimera, if we need (but we don't actually) and measure the distance between these two atoms. If there is a disulfide bond between them, the distance should be 2.05A.
To check the distance we need Shift+Control+left mouse to select two atoms between we want to measure distance.
Tools -> Structure analysis -> Distance.
You will notice the distance from the two sulfurs is about 2.03 angstroms.
I. Receptor Preparation
First step is to prepare the receptor. To do this open up the original pdb of the molecule. To prepare the receptor, you need to cut off the covalent linkers to generate the receptor.
Tools -> Structure editing -> Build Structure -> Modify structure - Element S, Bonds 2, Geometry tetrahedral.
The newly formed receptor is now with a complete cysteine.
Now add hydrogens and charge the protein. To make the cysteine neutral, protons will be added into the system. Protonate with the latest AMBER force field, currently AMBER14SB.
Tools -> Structure editing -> AddH. Tools -> Structure editing -> AddCharge.
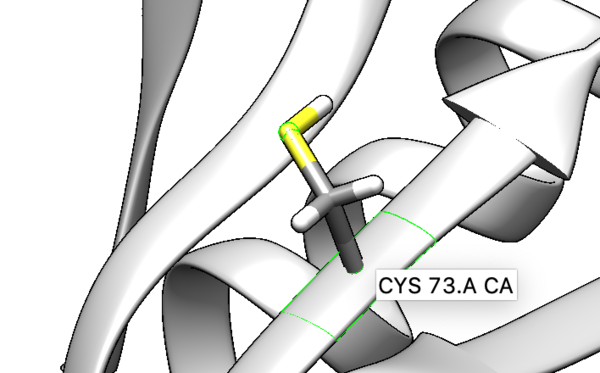
To prepare the receptor for making a grid, delete the side chain of the covalently bonded cysteine residue Cys73. When deleting the side chain of the residue make sure to delete S and the beta carbon, but leave the alpha carbon. That is the receptor that will be used for dock grid and docking.
II. Ligand Preparation
Open of the original structure rcsb104645_model1.pdb. Delete all the protein leaving only the ligand covalently bonded too the cystine, leave the cystine attached to it. The ligand should be binded to the side chain residue all the way up to the beta carbon.
Chose (holding Control + z) Ligand and Cys -> Invert -> Actions -> Atoms -> delete.
Make Cys73 to be visible in all atom mode.
Chose Cys73 (holding Control + z) -> atom/bond -> show.
Then we create a disulfide bond between S in the ligand and the residue. The command bellow will create a disulfide covalent bond:
Chose S in the ligand and in the Cys73 (control +z). Preferences -> Command line -> type "bond sel". It will create a disulfide covalent bond.
Protonate the modified ligand.
Tools -> Structure editing -> AddH
Look at N at the backbone on Cys73 residue. There is only one H attached to N after adding hydrogens, but need to be two. So add one more H to N at the Cys aminoacid backbone. To do so chose N (holding Control +z) and go to
Tools -> Structure editing -> Built structure - Modify Structure - Element N - Bonds 3 - Geometry trigonal.
Next we charge the ligand.
Tools -> Structure editing -> AddCharge -Net charge +1 - with AM1BCC.
Then delete all of the hydrogens off the beta-carbon.
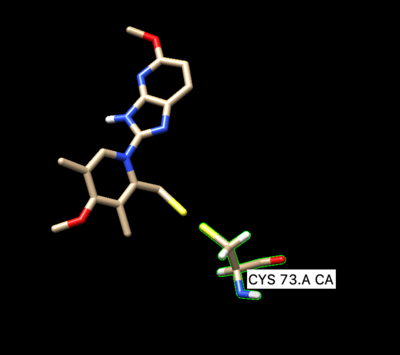
Next we need to modify the beta carbon to D2, and modify the side chain sulfur atom to D1. To do so save prepared ligand in mol2 format and open it with a text editing tool.
Then manually change the atom name of the beta carbon to D2 and change the atom type from C.3 to Du. Change the atom name of the gamma sulfur connected to the beta carbon to D1 and changed the atom type to Du as seen below:
@<TRIPOS>ATOM
1 D2 -24.0727 34.9285 -36.1952 Du 1 LIG -0.0660
2 D1 -25.1580 36.1840 -35.7130 Du 1 LIG -0.1111
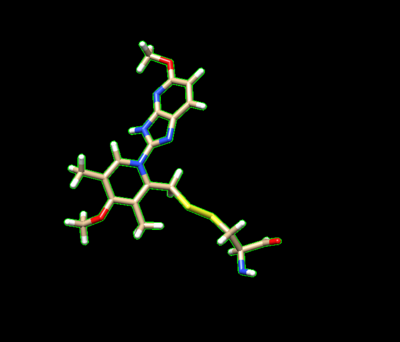
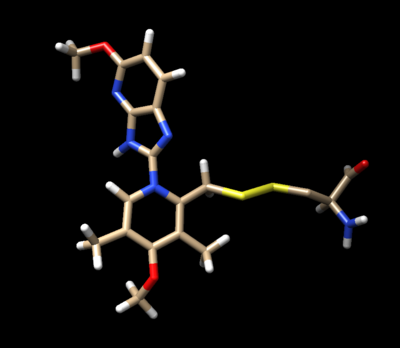
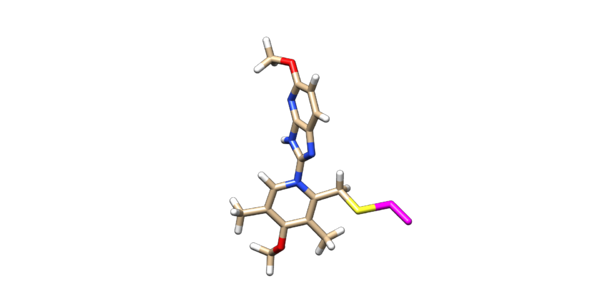
The ligand should now look like this in chimera:
This is the end of the ligand preparation.
III. Preparing the sphere files for orienting
We need to prepare two types of spheres: 1) spheres for box generating; 2) spheres for orienting the ligand.
The only spheres that will be orientated will be the alpha carbon, beta carbon, and the sulfur on the cysteine sidechain. Open the mol2 file with prepared ligand. Isolate the sulfur, alpha and beta carbons and delete everything else.
Once that is isolated save it as a pdb or as a mol2. Create file mol2_to_sph.py and write the following into it:
import sys, math, mol2 import os.path import cmath from math import sqrt ## written by Trent E. Balius and editted by Lauren Prentis ## Sept 2019 ## converts a mol2 into a sph for covalent
def write_sph_file(filename,mol):
##dt = mol2.convert_sybyl_to_dock(mol) # get dock atom types.
vdw_dict = 0.5
outsph = open(filename, 'w')
print (len(mol.atom_list))
outsph.write("DOCK spheres generated from ligand heavy atoms\n")
outsph.write("cluster 1 number of spheres in cluster %d\n" % len(mol.atom_list))
for i in range(len(mol.atom_list)):
radius = 0.5
outsph.write("%5d%10.5f%10.5f%10.5f%8.3f%5d 0 0\n" %
(i+1,round(mol.atom_list[i].X,3),
round(mol.atom_list[i].Y,3),
round(mol.atom_list[i].Z,3),radius,i+1) )
outsph.close()
return
def remove_hydrogens(m):
atom_list = []
bond_list = []
residue_list = {}
# Retain only heavy atoms in atom_list
num_hvy_atoms = 0
for i in range(len(m.atom_list)):
if (m.atom_list[i].heavy_atom):
atom_list.append(m.atom_list[i])
num_hvy_atoms+=1
# Retain only bonds containing heavy atoms
for bond_id in range(len(m.bond_list)):
retain_bond = True
for atom_id in range(len(m.atom_list)):
if (m.atom_list[atom_id].heavy_atom):
continue
# Atoms down here are always hydrogen
if (m.bond_list[bond_id].a1_num == m.atom_list[atom_id].num):
retain_bond = False
if (m.bond_list[bond_id].a2_num == m.atom_list[atom_id].num):
retain_bond = False
if (retain_bond):
bond_list.append(m.bond_list[bond_id])
# Assuming that residue list does not change
data = Mol(m.header,m.name,atom_list,bond_list,m.residue_list)
return data
def main():
if len(sys.argv) !=3:
print("Not enough inputs")
name_of_mol2 = sys.argv[1]
name_of_sph = sys.argv[2]
mol = mol2.remove_hydrogens(mol2.read_Mol2_file(name_of_mol2)[0])
write_sph_file(name_of_sph,mol)
main()
Use a code below to convert this pdb or mol2 into a sphere file.
python mol2_to_sph.py lig_abs.mol2 lig_abs_spheres.sph
Now new spheres will be generated for the box. To accomplish this spheres will be generated ligand file. A sphere file will be made using the coordinates from the ligand file of the heavy atom. Now that the spheres are generated twice once for orientating and the other for the dock/grid. The box will then be generated, once the box is made the grid can be populated in the box. Now that the grid, the ligand, and spheres meant of orientating. You can begin covalent docking.