2018 DOCK tutorial 2 with PDBID 1C87
For additional Rizzo Lab tutorials see DOCK Tutorials. Use this link Wiki Formatting as a reference for editing the wiki. This tutorial was developed collaboratively by the AMS 536 class of 2018, using DOCK v6.8 and it shows how to dock a ligand into a receptor.
Contents
I. Introduction
DOCK
DOCK is a molecular docking program used in drug discovery. It was developed by Irwin D. Kuntz, Jr. and colleagues at UCSF (see UCSF DOCK). This program, given a protein binding site and a small molecule, tries to predict the correct binding mode of the small molecule in the binding site, and the associated binding energy. Small molecules with highly favorable binding energies could be new drug leads. This makes DOCK a valuable drug discovery tool. DOCK is typically used to screen massive libraries of millions of compounds against a protein to isolate potential drug leads. These leads are then further studied, and could eventually result in a new, marketable drug. DOCK works well as a screening procedure for generating leads, but is not currently as useful for optimization of those leads.
DOCK 6 uses an incremental construction algorithm called anchor and grow. It is described by a three-step process:
- Rigid portion of ligand (anchor) is docked by geometric methods.
- Non-rigid segments added in layers; energy minimized.
- The resulting configurations are 'pruned' and energy re-minimized, yielding the docked configurations.
1C87
In this tutorial we will use PDB code 1C87, which is the crystal structure of protein tyrosine phosphatase 1B complexed with 2-(oxalyl-amino-4,7-dihydro-5H-thieno[2,3-C]pyran-3-carboxylic acid.
Organizing Directories
We are going to create and organize directories so it would be easier for us to find or identify files in each directory.
~/gpfs/projects/AMS536/2018/
/001.files/
/002.spheres/
/003.box/
/004.grid/
/005.dock/
/006.virtual-screen/
|
II. Preparing the Receptor and Ligand
Download the PDB File (1C87)
We are going to the PDB website (https://www.rcsb.org/) to download 1C87.pdb file and transfer this pdb file to your directory. First, open Chimera and load 1C87.pdb file. Remove the ligand and save the receptor in mol2.format. "mol2" format shows types of bonds whether it is single or double bond. After saving the receptor with no H (noh_receptor_1c87.mol2) and the receptor with H (rec_withH_1c87.mol2), load the original 1c87.pdb again. Remove the receptor and water, and save the ligand with H (lig_withH_1c87.mol2) and ligand with H and charge (lig_withH_charge_1c87.mol2). In order to add charge on ligand, go to Tools and Structure Editing and save the files. (NOTE: when you add H on ligand, make the charge -1. Check the article that is related to pdb file to decide whether it should be pronated or depronated.)
Tools -> Structure Editing -> Add H Tools -> Structure Editing-> Add Charge Select AMBER ff99SB for Standard Residues, and add a charge of -1. File -> Save Mol2... -> lig_withH_charge.mol2
Lastly, open the receptor file and click Surface Editing and write DMS file. Make sure to save all files. So far, we will have these files ready.
| <pre>
1c87.pdb lig_withH_charge_1c87.mol2 noh_receptor_1c87.mol2 lig_withH_1c87.mol2 noh_lig_1c87.mol2 rec_withH_1c87.mol2 |
After saving these two files with H, transfer files into 001.files directory:
scp -r ./*1c87* username@login.seawulf.stonybrook.edu:/gpfs/projects/AMS536/2018/your_directory_name/001.files
III. Generating Receptor Surface and Spheres
Generating the Receptor Surface
Make sure sphere directory is created and open the directory:
mkdir 002.surface cd 002.surface
Open Chimera and load the receptor surface:
Load the receptor (noh_receptor_1c87.mol2) file, which is located in the 001.files directory. Go to Action -> Surface -> Show and it will show the surface receptor. Next, save a DMS file as noh_surface_rec.dms by clicking Tools -> Structure editing -> Write DMS.
Creating Spheres We are going to create spheres by using the Sphgen program, which is a sphere generation program in Dock.
1. Create an input file
vi INSPH
Copy this following script in INSPH input file.
./noh_surface_rec.dms R X 0.0 4.0 1.4 1c87.spheres.sph
The first line ./noh_surface_rec.dms specifies the input file. R means that spheres generated will be outside of the receptor surface. X specifies all the points will be used. 0.0 is the distance in angstrom and it will avoid steric clashes. 4.0 is the maximum surface radius of the spheres and 1.4 is the minimum radius in angstroms.The last line 1c87.spheres.sph creates the file that has clustered spheres.
2. Run the Sphgen program
sphgen -i INSPH -o OUTSPH
-i chooses input file and -o writes toutput file.
3. Open Chimera to visualize the generated spheres Load 1c87.recep.sph file.
Now, we are going to select the spheres that is close to the native ligand molecule, which would be within 8.0 angstroms of the ligand.
sphere_selector 1c87.recep.sph ../001.files/lig_withH_charge_1c87.mol2 8.0
1c87.recep.pdbwill be created and load this file in Chimera.
IV. Generating Box and Grid
Enter 003.box-grid directory. Create input showbox.in:
Y 8.0 ../002.spheres/selected_spheres.sph 1 1c87.box.pdb
The first line Y creates a box (grid). 8.0 is the box length in angstroms. 1 means spheres. The last line, 1c87.box.pdb, outputs the box to the file.
Use this input type:
showbox < showbox.in
Now we can visualize the box in Chimera. Load 1c87.box.pdb.
Then, we will compute the energy grid. Create grid.in file
vi grid.in
Save the output in the file grid.out.
grid -i grid.in > grid.out
Answer the questions:
compute_grids yes grid_spacing 0.4 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4 bump_filter yes bump_overlap 0.75 receptor_file ../001.files/lig_withH_charge_1c87.mol2 box_file ../003.box/1c87.box.pdb vdw_definition_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
Once completed to answer the questions, it will create two output files: grid.bmp and grid.nrg
V. Docking a Single Molecule for Pose Reproduction
Minimization
Create min.in file and run dock with this input file.
touch min.in ../../../zzz.programs/dock6/bin/dock6 -i min.in
Answer the questions like we did above:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../001.files/lig_withH_charge_1c87.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../001.files/lig_withH_charge_1c87.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary yes continuous_score_secondary no cont_score_rec_filename ../001.files/lig_withH_charge_1c87.mol2 cont_score_att_exp 6 cont_score_rep_exp 12 cont_score_rep_rad_scale 1 cont_score_use_dist_dep_dielectric yes cont_score_dielectric 4.0 cont_score_vdw_scale 1 cont_score_es_scale 1 footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes atom_model all vdw_defn_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /opt/AMS536/dock6/parameters/flex.defn flex_drive_file /opt/AMS536/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 3pgl.min write_orientations no num_scored_conformers 1 rank_ligands no
Now, 1c87.min_scored.mol2 will be produced. We can compare minimized ligand structure with unminimized ligand structure.
Rigid Docking
We are going to run rigid docking. In order to run dock, we will have to create rigid.in file.
touch rigid.in ../../../zzz.programs/dock6/bin/dock6 -i rigid.in
Answer the questions:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../001.files/lig_withH_charge_1c87.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../001.files/lig_withH_charge_1c87.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../004.grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 1c87.rigid_restraint write_orientations no num_scored_conformers 40 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
This will create 1c87.rigid_restraint_scored.mol2 in 005.dock directory.
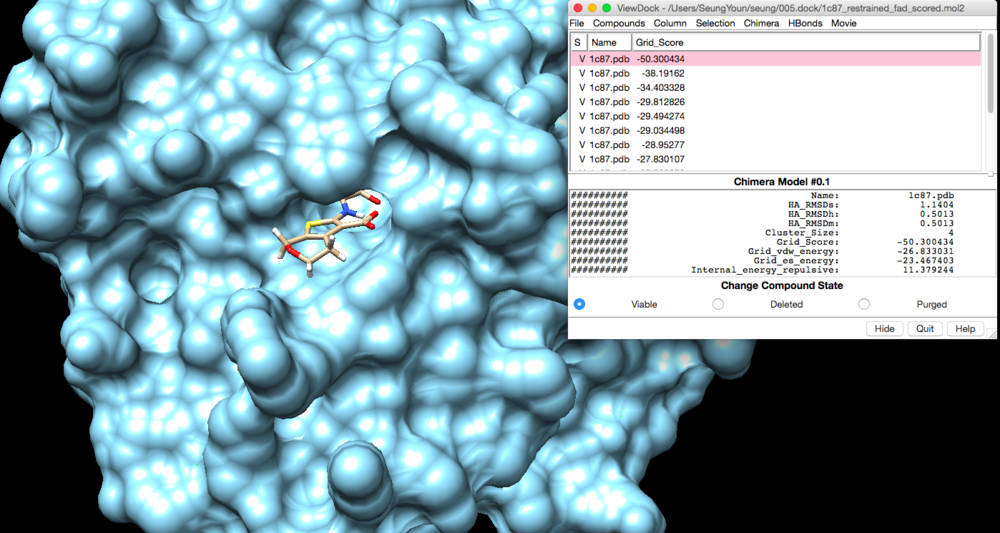
We can view rigid docking output file in CHIMERA.
Tools --> Surface/Binding Analysis -> ViewDock Select the output file -> select Dock 4,5, or 6
We can view the lowest grid score (the best ligand that fits in the pocket) , which is what we want, by clicking Column -> Show -> Grid_Score.
Flexible Docking
In order to run flexible docking, we will also create flex.in file. In this case, ligand and angles are flexible (NOTE: orient_ligand is set to yes).
touch flex.in ../../../zzz.programs/dock6/bin/dock6 -i flex.in
When prompted, answer the questions as follows
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../001.files/lig_withH_charge_1c87.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../001.files/lig_withH_charge_1c87.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../004.grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 1c87_flex write_orientations no num_scored_conformers 40 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
The output file is 3pgl.flex_scored.mol2, and is saved in the directory from where you ran dock. Then we can visualize the flexible docking in Chimera:
Fixed Anchor Docking
We are going to run fixed anchor docking. Create fad.in.The ligand is fixed in the active site and angles are flexible. (NOTE: orient_ligand is set to no).
touch fad.in ../../../zzz.programs/dock6/bin/dock6 -i fad.in
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../001.files/lig_withH_charge_1c87.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../001.files/lig_withH_charge_1c87.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../004.grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 1c87_fad write_orientations no num_scored_conformers 40 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
Again, we can view fixed anchor docking in Chimera and find the lowest grid score value.
Now we have created output files for rigid, flexible and fixed anchor docking.
VI. Virtual Screening
We will create a virtual-screen.in file in 006.virtual-screen that looks like the following:
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file small_ligand_library.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../001.files/lig_withH_charge_1c87.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../004.grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 1c87_virtualscreen write_orientations no num_scored_conformers 1 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
To submit the job for docking, create submit.sh file.
vi submit.sh
Write down the following script in submit.sh file.
#! /bin/bash #PBS -l walltime=48:00:00 #PBS -l nodes=1:ppn=24 #PBS -q long #PBS -N 1c87_rigid #PBS -V cd $PBS_O_WORKDIR dock6 -i rigid.in -o 1c87_rigid_docking.out
After creating this script, submit the job.
qsub submit.sh
We can view whether the job runs or does not run (if there is an error):
qstat -u username